Bacterial Diversity in Some Local Fishes at Central Fish Market of Jeddah, Saudi Arabia
2 Department of Biology, King Abdulaziz University, Jeddah, Saudi Arabia, Email: magdammali@hotmail.com
Published: 30-Sep-2021
Citation: Beyari EA, et al. Bacterial Diversity in Some Local Fishes at Central Fish Market of Jeddah, Saudi Arabia. Ann Med Health Sci Res. 2021;11:33-38.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Jeddah is the second largest city in Saudi Arabia and located on the Red Sea coast (western seaboard). It has the largest seaport along the Red Sea coast. Rapid industrialization and urbanization in the Jeddah area have raised concerns over potential impacts on public health and the environment. Contaminated effluents through point and nonpoint sources may enter the coastal waters of Jeddah and eventually find their way into the body tissues of edible fish. These contaminants can cause health problems. Fish contamination with bacterial pathogens could cause severe foodborne illness and even death in children. This presents as a significant concern as fish is a major dietary protein source for people in the area. Despite significant fish consumption, scientific investigation of marine microbial contamination, particularly on bacterial pathogens in seafood generally remains scarce in Saudi Arabia. Monitoring the microbial contaminants in commonly consumed fish will thus help in safeguarding the public from any potential adverse risks. The present study aims to isolate several species of bacteria from specific kinds of fish collected from Jeddah central fish market and identify them molecularly.
Keywords
Mac conkey; Foodborne; Bacteria; Pathogens; Jeddah
Introduction
Fishes are known for their exceptional health benefits, particularly against cardiovascular diseases and for infant brain development due to their long-chain Polyunsaturated Fatty Acids (PUFAs) content. [1] Despite such nutritional benefits and culinary fondness, infection of fishes by pathogenic microorganisms and other contaminants is a major concern for seafood consumers. Fish contaminated with bacterial pathogens could cause severe foodborne illness and offset the health benefits of PUFAs. [2] In the developing world, foodborne infection leads to the death of many children, as well as resulting in diarrheal disease which can have long-term effects on children’s growth as well as on their physical and noes is development and it also heavily affects the healthcare systems. [3] According to Clarence et al. food borne diseases are diseases resulting from ingestion of bacteria, toxins and cells produced by microorganisms present in food. The intensity of the signs and symptoms may vary with the amount of contaminated food ingested and susceptibility of the individuals to the toxin. [4] Bacterial abundance in fish species generally varies based on environmental and biological factors. [5] The primary sources of microbial pathogens in fish are anthropogenic activities that generate point and non-point pollution in coastal waters. There are also naturally occurring waterborne pathogens like vibrio that could cause human illness by way of food consumption. Hence, the monitoring of bacterial pathogens potentially provides early warning to safeguard seafood consumers from the threats of contamination.
Jeddah is the second largest city in Saudi Arabia and located on the Red Sea coast (western seaboard). It has the largest seaport along the Red Sea coast, with a total throughput of 4700 vessels (56% container ships) and 52 million metric tons of cargo (69% discharged, 31% loaded) in 2011. [6] Rapid industrialization and urbanization in the Jeddah area have raised concerns over potential impacts on public health and the environment. Contaminated effluents through point and non-point sources may enter the coastal waters of Jeddah and eventually find their way into the body tissues of edible fish. This presents as a significant concern as fish is a major dietary protein source for both Saudis and expatriates in the area. Despite significant fish consumption, scientific investigation of marine microbial contamination, particularly on bacterial pathogens in seafood, generally remains frightened in Saudi Arabia. Monitoring the microbial contaminants in commonly consumed fish will thus help in safeguarding the public from any potential adverse risks. This paper presents our findings on the bacterial contamination in fishes collected from the central fish market in Jeddah. Bacterial abundance in different body parts (skin, gills, gut, and muscle) of fishes was determined by culture dependent method using three selective isolation media. In addition, the bacterial species were identified by gene sequencing technique.
Materials and Methods
Sampling
Fish samples were collected from several stores from Jeddah central fish market [Figure 1]. Five species of more commonly consumed fish were selected based on a questionnaire in Jeddah city, Table 1 illustrate types of fish included in this study. Fish samples were collected individually in a clean polyethylene bag and kept in ice box and transferred immediately to the laboratory in refrigerated condition. [7]
| Table 1: The list of fish utilized in this study, scientific name, family, common english name and common Arabic name. | ||||
|---|---|---|---|---|
| S.no | Scientific name | Family | Common english name | Common Arabic name |
| 1 | Epinephelus areolatus | Serranidae | Yellow spotted rockrod | Hamoor |
| 2 | Plectropomus leopardus | Serranidae | Coral trout | Najil |
| 3 | Lethrinus nebulosus | Lethrinidae | Spangled emperor | Shaour |
| 4 | Scarus frenatus | Scaridae | Bridled parrot fish | Hareed |
| 5 | Carangoides orthogrammus | Carangidae | Island travelly | Bayad |
Sampling processing
Samples of gills, skin, muscle, and gut, of fish were aseptically obtained by dissection and then prepared for plating following Andrews et al. All the dissecting apparatuses, such as scalpel, forceps, scissors, knives, mortar, pestle, and glassware, were sterilized with 100% ethanol and kept in hot air oven at 180°C for 6 h prior to dissection. Gills were removed from both sides of the fish head and homogenized to a composite sample. Skin was removed from the side of fish behind the dorsal fin after removing the scales with a sterilized knife. [8] Muscle (flesh) samples were collected through incision on the same side and section of the fish body. An opening into the body cavity was carefully made with a sterile scalpel to remove a small portion of the gut. All samples were taken in the same order from the same body sections uniformly across fish species. After dissection, 1 g of each dissected organs were separately homogenized using a mortar and pestle. The homogenized tissues were diluted in sterilized water then plated on different selective media for the estimation of bacterial counts and isolation of different bacterial species.
Enumeration
Three selective isolation media, Mac conkey’s agar (Mac), Eosin Methylene Blue agar (EMB) and Thiosulfate Citrate Bile Salts agar (TCBS), were used for isolation. [1,9] Isolation media were prepared by suspending the (Conda, Spain) Mac (47 g/l), (Conda, Spain) EMB (36 g/l) and (Himedia, Mumbai) TCBS (89.08 g/L) in sterile distilled water. Media were completely dissolved by heating on a hot plate and sterilized by autoclaving at 15 lbs pressure and 121°C temperature for 15 min. TCBS media was not autoclaved but heated until boiling after suspending them in previously sterilized distilled water. Media were then cooled up to 50 °C and aseptically poured into Petri plates under sterile conditions. Enumeration of bacterial load in fish tissues were done by streak plate method. The plates were incubated in inverted position at 35°C ± 2°C for 18 hrs-24 hrs. Bacterial colonies were counted and expressed in Colony Forming Units (CFU) per gram of given sample. [1,10] Figure 2 showed central fish market in Jeddah city.
Bacterial molecular identification
Bacterial groups isolated from the samples were morphologically categorized based on their colony, shape, size, color and Gram stain. Dominant bacterial groups were sub-cultured, and DNA were extracted according to Wright et al. Genomic DNA extraction was performed at King Fahd medical research centre, King Abdulaziz university.
Phylogenetic analysis of 16S rRNA sequence
The sequencing of the PCR amplicon of the selected isolates was done by Macrogen, Inc (Seoul, Korea),by using 16S rRNA universal forward and reverse primers, 785F 5'GGATTAGATACCCTGGTA 3'and 907R 3' CCGTCAATTCMTTTRAGTTT 5'. The 16S rRNA was amplified using a thermal cycler (Applied Biosystems Veriti™ 96-Well Thermal Cycler). PCR conditions consisted of an initial denaturation at 95°C for 5 min; followed by 35 cycles of denaturation (94°C for 40 sec), annealing (58°C for 40 sec), and extension (72°C for 1 min), and a final extension at 72 ℃ for 10 min. The amplification product was examined by 1% agarose gel electrophoresis and the gel was visualized by Ultra-Violet Product (UVP BioSpectrum ® Imaging System). The 16S rRNA sequence was compared with available sequence data in the GenBank at the National Center of Biotechnology Information (NCBI) using the BLAST program. [11,12]
Results
Bacterial species isolated from fish
A total of 17 different bacterial genus and 32 different species were molecularly identified and named according to: fish type isolated from, media type, part of the fish isolated from and number of the isolate. Approximately 91% of the identified species are Gram negative. Aeromonas, Pseudomonas, Psychrobacter, Alcaligens were the dominant isolated species. Table 2 illustrated some of the isolated species. Some isolated species are pathogenic for humans such as, Rahnella aquatilis, Proteus vulgaris, Klebsiella quasipneumoniae, Yersinia enterocolitica, Pseudomonas lundensis, Pseudomonas oryzihabitans, Psychrobacter phenylpyruvicus, Psychrobacter sanguinis and some of them considered as strong pathogens or opportunistic human pathogen, for example, Alcaligenes faecalis and Pseudomonas putida.
| Table 2: List of some bacterial species isolated from studied fish and their accession number of the most related strains in NCBI gen bank. | ||
|---|---|---|
| Bacterial isolates | Name and accession number of the most related strain in NCBI gen bank | |
| BE25 | NR_025337.1 | Rahnella aquatilis |
| RT11 | NR_043721.1 | Salinicoccus salsiraiae |
| BT11 | NR_115878.1 | Proteus vulgaris |
| RE13 | NR_134062.1 | Klebsiella quasipneumoniae |
| BM14 | NR_042415.1 | Providencia vermicola |
| BE14 | NR_043324.1 | Aeromonas salmonicida |
| RE41 | NR_117303.1 | Aeromonas diversa |
| SE11 | NR_025107.1 | Comamonas koreensis |
| BT31 | NR_116596.1 | Jeotgalicoccus nanhaiensis |
| BE31 | NR_104903.1 | Yersinia enterocolitica |
| RM23 | KX186958.1 | Pseudomonas lundensis |
| RM22 | NR_028929.1 | Pseudomonas kilonensis |
| RM43 | FJ430676.1 | Pseudomonas putida |
| HM34 | KT825519.1 | Pseudomonas japonica |
| SE32 | JQ762269.4 | Pseudomonas songnenensis |
| HE11 | NR_116299.1 | Pseudomonas brassicacearum |
| NT12 | NR_114630.1 | Vibrio parahaemolyticus |
| BE42 | NR_113606.1 | Alcaligenes faecalis |
| HM21 | NR_028966.1 | Psychrobacter faecalis |
| SM22 | NR_114038.1 | Psychrobacter phenylpyruvicus |
Bacterial counts on selective media
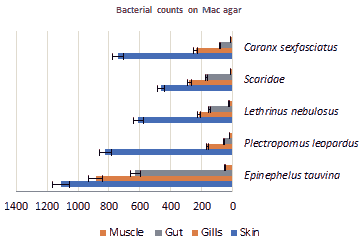
Counts on mac conkey agar: Mean bacterial counts in body parts on Mac agar are shown in Figure 3, the highest load of bacteria is in skin followed by gills then gut and finally muscles in all the studied fish. Epinephelus tauvina played the highest load of bacterial number in all four body parts, which are: skin 11.0 × 102 CFU/g, gills 8.87 × 102 CFU/g, gut 6.30 × 102 CFU/ g and finally muscles 0.5 × 102 CFU/g.
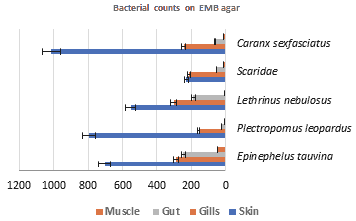
Counts on EMB agar: Mean bacterial counts in body parts on EMB agar are illustrated in Figure 4; the highest load of bacteria is in skin followed by gills then gut and finally muscles in all the studied fish. Epinephelus tauvina displayed the highest load of bacterial number in both gut and muscles, that are 274, 47 CFU/g respectively. Caranxsex fasciatus showed the highest number in skin which is 1013 CFU/g. Lethrinus nebulosus displayed the highest number in gillsthat is 303 CFU/g.
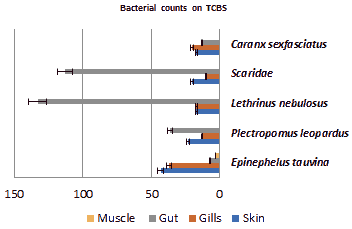
Counts on TCBS agar: Counts in TCBS media displayed the lowest bacterial load comparing to both other media. Epinephelus tauvina is played the highest load in skin that about 43 CFU/g. Plectropomus leopardus, Scaridae and Lethrinus nebulosus showed the highest load in gut compared to other body parts, which are 36 CFU/g, 113 CFU/g and 133 CFU/g respectively. Caranxsex fasciatus displayed highest load in gills part that is 20 CFU/g. Figure 5 illustrated the mean bacterial counts in body parts on TCBS media.
Discussion
Microorganisms in the marine environment have both beneficial and harmful functions. They execute biogeochemical cycles that are a critical process in marine environments. [13] Microbial contaminants mainly consist of pathogenic groups of bacteria, viruses, and parasites. The presence of different bacteria species including human pathogenic bacteria in fish can be linked to direct contact with a contaminated water environment and ingestion of bacteria from sediments or contaminated feed. Thus, bacteria detected in fish reflect the condition and safety of aquatic environments. [14]
Fish may be a vehicle for pathogenic bacteria naturally occurring in aquatic environments, referred to as indigenous or derived from polluted waters and/or from post capture contamination. [15] The indigenous bacteria that occur naturally in the marine environment which, when consumed in seafood in large enough numbers, will cause illness in humans. This primarily relates to the Vibrios. Some species of the genus Aeromonas are considered to some to possibly cause gastro-enteritis in humans, and these may also be present naturally in the marine or more especially, the estuarine environment. Spores of type F Clostridiumbotulinum are found widely in marine sediments and the intestinal tract of fish and shellfish and, if seafood is stored under conditions (principally in the absence of oxygen) that allows the spores to germinate and the bacteria to multiply, toxin may be formed in the seafood and then cause botulism in humans when the food is consumed. [16]
Globally, the sea has been used as a dumping ground for sewage and other waste products. Sewage water contains microorganisms and several human-made-chemical compounds. The possible sources of fish bacteria are likely to come from the skin of the animal from which the fish was obtained. This may arise from contamination with human feces from sewage discharges, boats, and ships or, in some countries, direct defecation into the marine environment or rivers and streams flowing into it. [16]
The outlined and discussed the hazards and challenges associated with handling fish during farming, capture and the environmental contaminants in seafood that may pose a risk to human health. [17] Potential sources of microbial contaminations are the equipment and items used for each operation that is performed until the final product is eaten. Boats, fish boxes, fishermen and workers clothing and hands, knives, cutting boards, display tables, ice, and the physical facilities themselves are all implicated. Retail cut could also result in greater microbial load because of the large amount of exposed surface area, more readily available water, nutrient, and greater oxygen penetration available. Hence retail cuts displayed are conducive for microbial growth and proliferation which leads to spoilage of the fish. [18]
In this study, about 32 bacterial species were obtained from different parts of five types of fish collected from central fish market located in Jeddah city, western Al-Bagdadiyah, Al Corniche Road, Jeddah. Several factors were considered for the collection and isolation of bacteria from fish. Fish samples were collected individually in a clean polyethylene bag and transferred immediately to the laboratory in refrigerated condition. Then fish samples were dissected, and bacteria were isolated on three selective media. The selective mac conkey agar is a differential culture medium for bacteria. It is designed to selectively isolate gram-negative and enteric (normally found in the intestinal tract) bacteria and differentiate them based on lactose fermentation Dominant species were sub cultured and morphologically identified. The most dominant species were isolated in this study are Aeromonas, Pseudomonas, Psychrobacter, Alcaligens. Moreover, Rahnella aquatilis was isolated in this study. Compared to Alikunhi et al. study, Rahnella aquatilis and Photobacterium damselae were among the dominant species isolated from fish muscle from Jeddah from 3 different areas based on 16S rRNA sequencing. Also, Aeromonas, Pseudomonas, Psychrobacter were isolated in Alikunhi et al. study. In contrast with study in Al Khartoum, Sudan, Enterobacteriaceae were isolated from gills, skin, muscles, and intestine randomly collected fishes from Al Kartoum fish market, the most dominants isolates were E. coli, Citrobacter spp., Enteriobacter spp., and Klebsiella spp. This together with the highly pathogenic Enterobacteriaceae including Salmonella spp., Shigella spp., Proteusspp., and Alcaligens spp. Potential pathogenic organisms were also among the isolates. On the other hand, Pseudomonasspp. was isolated from 62% of randomly collected fishes. [17] Compared to another research, fish spoilage bacteria Alcaligenes faecalis, Pseudomonas aeruginosa, Proteus vulgaris, Aeromonasspp. and Vibrio alginolyticus were isolated from fish from fish market in Kenya. [19]
According to Gulf Cooperation Council (GCC) and Yemen standardization organization, GCC Standardization Organization (GSO), the accepted limit of aerobic bacterial counts for fresh fish is between 5×102 and 5×105 CFU/g [Table 3]. In this study, the level of total viable bacterial count was accepted in all different fish kinds and in all four body parts in all selective media. The accepted bacterial counts in this study could be cause of good production practices among food processors and food vendors. It is expedient that good hygienic practices are observed at the market and revealed that the market follow the precautionary instructions that applied by Saudi ministry of municipal rural affairs. In contrast, study of Alikunhi et al. presented that Mugil cephalus from area 1 and fish market (Area 1 is a semi-enclosed lagoon lined with several storm water drainage outlets, a corniche, city road network, hotels, restaurants, a private harbour, and commercial buildings within a hundred meters from the shorefront) showing unaccepted bacterial loads for gills and guts on Mac, EMB and TCBS media. As opposed to additional study in retail fish market, Dammam, Saudi Arabia, results revealed that several fish samples exceeded the acceptable bacterial counts which indicate the urgent need required to improve the quality control and quality assurance systems of Dammam fish market. [5] Mean bacterial counts in all body parts on Mac and EMB agar gave the highest load of bacteria in skin followed by gills then gut and finally muscles in all studied fish. Epinephelus tauvina displayed the highest load of bacterial number on Mac in all four body parts. Epinephelus tauvina displayed the highest load of bacterial number on EMB in both gut and muscles. Scaridae and Lethrinus nebulosus showed the highest load in gut on TCBS media compared to other body parts. Bacterial abundance in fish species generally varies based on environmental and biological factors. This variation could be because some fishes are inherently more prone to contamination depending on the species, feeding pattern, age, size, harvest season, habitat characteristics, and geographical location. [5] Also, various factors including the season, part of the digestive tract of fish, and feeding type can affect the number of microorganisms detected. The minimum and maximum findings for specific bacteria were related to the changes of water temperature and were observed during winter and summer seasons. [21-26]
| Table 3: Accepted limits of bacterial counts in fish per gram or cm² according to GCC Standardization Organization (GSO), Gulf cooperation council and Yemen standardization organization. | |
|---|---|
| Type of bacteria | Accepted limits in CFU/g |
| Aerobic bacteria | 105 |
| Escherichia coli | 10 |
| Vibrio parahaemolyticus | 10 |
| Clostridium botulinum | 0 |
| Aeromonas spp. | 10² |
Conclusion
Bacterial contamination in commercial marine fishes from the Jeddah area was investigated by culture-dependent method and 16S rRNA gene sequencing of isolates. Results revealed differences in bacterial loads between species and fish parts. According to GCC Standardization Organization (GSO), all the bacterial counts in all body fish for all studied fish are considered accept. 16S rRNA sequences identified some pathogenic bacteria in the tested fish samples. Species such Rahnella aquatilis, Proteus vulgaris, Klebsiella quasipneumoniae are known human pathogens capable of causing foodborne illness with severe antibiotic resistance. Opportunistic pathogens such as Alcaligens faecalis, and Pseudomonas putida were also identified. The existence of these bacterial species in fish body samples are of concern. The present results indicate the potential risks of bacterial contamination of the fishes from the Jeddah area. Periodic monitoring, preferably including both microbial loads and species of concern, is thus important to keep track of any potential health risks to fish consumer.
References
- Alikunhi NM, Batang ZB, AlJahdali H, Aziz MA, Alsuwailem AM. Culture-dependent bacteria in commercial fishes: Qualitative assessment and molecular identification using 16S rRNA gene sequencing. Saudi J Bio Sci. 2017;24:1105-1116.
- Huss HH. Control of indigenous pathogenic bacteria in sea food. Food Control. 1997;8:91-98.
- Adak GK, Meakins SM, Yip H, Lopman BA, Obrien SJ. Disease risks from foods, England, and Wales, 1996-2000. Emerg Infect Dis. 2005;11:365-372.
- Clarence SY, Obinna CN, Shalom NC. Assessment of bacteriological quality of ready to eat food (meat pie) in Benin City metropolis, Nigeria. Afr J Microbiol Res. 2009;3:390-395.
- Novotny L, Dvorska L, Lorencova A, Beran V, Pavlik, I. Fish: A potential source of bacterial pathogens for human beings: A review. Vet Med. 2004;49:343-358.
- https://mawani.gov.sa/en-us/pages/default.aspx.
- Nayak DN, Savalia CV, Kalyani IH, Kumar R, Kshirsagar DP. Isolation, identification, and characterization of Listeria spp. from various animal origin foods. Vet World. 2015;8:695-701.
- Alharbi AH, Uddin N. Bacterial diversity of tilapia (oreochromis niloticus) cultured in brackish water in Saudi Arabia. Aquaculture. 2005;250:566-572.
- Yucel N, Balci S. Prevalence of Listeria, Aeromonas, and Vibrio species in fish used for human consumption in Turkey. J Food Prot. 2010;73:380-384.
- Ishida Y, Ahmed AM, Mahfouz NB, Kimura T, Elkhodery SA, Moawad AA, et al. Molecular analysis of antimicrobial resistance in gram-negative bacteria isolated from fish farms in Egypt. J Vet Med Sci. 2010;72:727-734.
- Momtaz H, Yadollahi S. Molecular characterization of Listeria mono cytogenes isolated from fresh seafood samples in Iran. Diagn Pathol. 2013;8:1-6.
- Yanez MA, Catalan V, Apraiz D, Figueras MJ, Martinezmurcia AJ. Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. Int J Syst Evol. 2003;53:75-883.
- Hewson I, Meyers MEJ, Fuhrman JA. Diversity and biogeography of bacterial assemblages in surface sediments across the San Pedro Basin, Southern California Borderlands. Environ Microbiol. 2007;9:923-933.
- Novoslavskij A, Terentjeva M, Eizenberga E, Valciņa O, Bartkevičs V, Berziņs A. Major food borne pathogens in fish and fish products: A review. Ann Microbiol. 2016;66:1-15.
- Herrera FC, Santos JA, Otero A, Garcialopez ML. Occurrence of foodborne pathogenic bacteria in retail pre-packaged portions of marine fish in Spain. J Appl Microbiol. 2006;100:527-536.
- https://archimer.ifremer.fr/doc/00066/17730/.
- Yagoub SO. Isolation of Enterobacteriaceae and Pseudomonas spp. from raw fish sold in fish market in Khartoum state. J Bacteriol Res. 2009;1:85-88.
- Antwiagyei P, Maalekuu B. Determination of microbial contamination in meat and fish products sold in the Kumasi metropolis (A case study of kumasi central market and the bantama market). Merit Res J Agric Sci Soil Sci. 2014;2:38-46.
- Binta GM, Tjaberg TB, Nyaga PN, Valland M. Market fish hygiene in Kenya. J Hyb. 1982;89:47-52.
- Alshabeeb SS, Ibrahim MM, Ramadhan GA. A comparative microbial quality assessment among fishes, prawns and cuttlefishes collected from dammam fish market. Int J Cur Microbiol. 2016;5:405-418.
- Diler O, Diler A. Quantitative and qualitative changes of the gastrointestinal micro flora of pike-perch (stizostedion luciopercal. 1758) in Egirdir lake. Turkish J Vet Anim Sci. 1998;22:325-328.
- Acharjee M, Ahmed I, Munshi SK, Noor R. Validation of γ-irradiation in controlling microorganisms in fish. Nutr Food Sci. 2014;44:258-266.
- Andrews WH, Hammack TS. Food sampling and preparation of sample homogenate. BAM, 2003;1-10.
- Hedberg CW, Levine WC, White KE, Carlson RH, Winsor DK, Cameron DN, et al. An international food borne outbreak of shigellosis associated with a commercial airline. J Med Appli. 1992;268:3208-3212.
- https://www.fda.gov/food/cfsan-risk-safety-assessments/quantitative-risk-assessment-public-health-impact-pathogenic-vibrio-parahaemolyticus-raw-oysters.
- Wright MH, Adelskov J, Greene AC. Bacterial DNA extraction using individual enzymes and phenol/chloroform separation. J Microbiol Biol Educ. 2017;18:112-117.




 The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.
The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.