Bacteriological Profile and Antibiotic Sensitivity Patterns in Clinical Isolates from the Out-Patient Departments of a Tertiary Hospital in Nigeria
2 Department of Microbiology, Irrua Specialist Teaching Hospital, Irrua, Edo state, Nigeria
3 Department of Biological Sciences, Ondo State University of Science and Technology, Edo State, Nigeria
Citation: Tobin Ekaete A, et al.Bacteriological Profile and Antibiotic Sensitivity Patterns in Clinical Isolates from the Out-Patient Departments of a Tertiary Hospital in Nigeria. Ann Med Health Sci Res. 2021;11:1453-1460.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Introduction: Antimicrobial resistance is a rising global public health threat. Knowledge on the circulating pathogens in a particular area and their antibiotic resistance profile is essential to direct clinicians on the rational antibiotic prescribing. The study was conducted to determine the microbial isolates and antibiotic susceptibility profiles of pathogens from a range of clinical samples in a tertiary hospital in Edo Central senatorial district in Edo state, Nigeria. Methods: The study was a retrospective analysis of microbiological isolates from clinical specimens collected between January 2016 and December 2019, using standard techniques from out-patient clinic attendees. Chi-square test was used to compare the association of type of bacterial isolates with patients’ sex, with the level of significance p set as <0.05. Prevalence rates of bacterial isolates and Resistance rates were calculated for each antibiotic used in microbiological culture. Results: Out of 3,247 clinical specimens processed, 994 (30.6%) showed microbial growth with 436 (43.9%) as gram-positive and 558 (56.1%) gram-negative bacterial isolates. Escherichia coli made up 286 (28.8%) of all isolates. Resistance to common antibiotics including cotrimoxazole, Tetracycline, Erythromycin and Cloxacillin were high for both microbial groups. Sensitivity to carbapenems, nitrofurantoin, and cephalosporins was high for gram-negative bacteria. Gram-positive bacteria exhibited high sensitivity to carbapenems and cephalosporins. Conclusion: High rates of resistance to common antibiotics were observed for gram-positive and gram-negative isolates. Hospital pharmacies and treatment guidelines should be made to reflect the current patterns of resistance to available antibiotics.
Keywords
Antibiotic; Bacterial isolates; Out-patients; Resistance
Introduction
Bacterial infections continue to contribute significantly to the overall morbidity and mortality from infectious diseases in developing countries despite the availability of antibiotics. [1]
The rising threat of Antimicrobial Resistance (AMR) described as a global public health challenge of the 21st century, increases the frailty of human existence by increasing vulnerability to bacterial infections that were hitherto treatable with available antibiotics. [2] Antibiotic-resistant bacteria are difficult to treat, limit therapeutic options, prolong hospitalization and require higher doses and probably drugs with higher tendencies for toxicity. [3] The slow progress with the development of new antibiotics to replace the first-line drugs to which bacteria have become resistant further compounds the problem. In the past 50 years, only two new classes of antibacterial drugs have been developed and introduced into clinical practice. [4] Even when a promising drug or vaccine exists, the high cost of production and length of time between regulatory approval and deployment reduces its availability. [5,6] Several studies in developed and developing countries describe the rising patterns of bacterial resistance. In a study of uropathogens in Western Nigeria, 35.8% of urine samples yielded bacterial growth with the majority, 25.6% identified as Escherichia coli. All were found to be resistant to at least 3 commonly used drugs. [7] In another study, Nmema et al. investigated the antibiotic susceptibilities and resistance mechanisms of Pseudomonas aeruginosa isolated from clinical samples collected from patients in a tertiary hospital in Lagos, Nigeria. Half of the isolates were multidrug-resistant, and 40% were resistant to imipenem and meropenem, a group of antibiotics considered as the last line for Gram-negative infections. [8]
The increasing occurrence of resistant bacterial pathogens necessitates that patterns of infection and antibiogram profile of community-acquired bacterial infections are reviewed periodically, and the information used to guide the development of local treatment guidelines and hospital antibiotic policies that will guide the use of antibiotics. [9] This is also vital for empirical treatment of patients, a common practice where Medical Microbiology laboratory diagnostic capacity is limited. [10]
The present study was carried out to investigate the bacteria and their prevalence in clinical samples submitted for microbiological analysis from the out-patient clinics at a tertiary teaching hospital in Edo State, Nigeria, determine the antimicrobial susceptibility pattern of isolates and describe the patients’ age and sex distribution for the isolates.
Materials and Methods
Study area
The study was carried out in a 375-beds tertiary teaching hospital in rural Edo state, South-south Nigeria. Located along the Benin-Abuja expressway in Irrua, the headquarters of Esan Central Local Government Area (LGA) in Edo Central Senatorial District, the hospital serves the state and neighbouring states of Delta, Kogi and Ondo. The hospital is one of 2 tertiary Health Institutions in the state and provides a comprehensive spectrum of clinical, promotive, preventive and rehabilitative services to the people in Edo state, particularly Esan central senatorial district, and neighbouring states.
Study design
The study was a retrospective cross-sectional analysis of Medical Microbiology laboratory test results of samples collected between January 2016 and December 2019. Bacteriological data over this period were retrieved from the laboratory result logbook using a pre-designed data extraction sheet. Age and sex of the patient, clinic name, specimen type, bacteriological culture and antibiotic susceptibility profile were documented.
Sample collection and characterization
Specimens were collected from all from patients attending the out-patient clinics of the hospital over the study period. Specimens were collected using standard methods of specimen collection and in line with standard operating procedures in use in the laboratory. [11] They were delivered to the laboratory within one hour of collection and analysis started the same day. Inoculated agar plates were incubated at 37°C for 16–48 hours. Culture and identification of bacteria followed Standard Operation Procedures of the Medical Microbiology Department. Culture media used for isolation of the microorganisms included Blood agar, MacConkey agar, CLED and Chocolate agar. Presumptive identification was based on Gram staining reaction and colony characteristics. Discrete colonies were sub-cultured for 24 hours at 37°C on Nutrient agar to purify the isolates. Confirmatory tests were based on the enzymatic and biochemical properties of the pure colonies. Gram-negative rods were identified by biochemical tests including oxidase, motility, indole, citrate, lysine decarboxylase, urease, and Triple Sugar Iron (TSI). Gram-positive cocci were identified based on their Gram reaction, catalase, and coagulase test results. All procedures were carried out in line with standard microbiological methods. [12,13] Patients’ age and sex were also collected.
Antibiotic agents
Antibiotic discs containing Ceftazidime CAZ (30 μg), Cefuroxime CRX (30 μg), Cefixime CMX (5 μg), Gentamicin GEN(10 μg), Ofloxacin OFL (5 μg), Amoxicillin–clavulanic acid AUG (30 μg), Nitrofurantoin NIT (300 μg), Cloxacillin CXC (5 μg), Ceftriaxone CTR (30 μg) Tetracycline TE (30 μg), Streptomycin S (30 μg), Clindamycin DA (30 μg), Erythromycin ERY(5 μg), NalidixicacidNA (30 μg), Ceftazidime CAZ (30 μg), Chloramphenicol (30 μg), Amoxycillin (10 μg), Cotrimoxazole (25 μg), Azithromycin AZM (15 μg), Retapamulin Ciprofloxacin CIP (30 μg) and Meropenem (10 μg) were chosen based on local utilization patterns, obtained from Oxoid Laboratories (Oxoid, UK) and used as instructed by the manufacturer.
Antibiotic susceptibility testing
Antimicrobial susceptibility testing was carried out using the Kirby-Bauer disc diffusion method and was reported in conformity with the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2017). After adjustment to 0.5 McFarland, a standard inoculum of each isolate was swabbed on a Mueller-Hinton agar plate using a sterile cotton swab stick. Using sterile forceps, the antibiotic discs were placed aseptically on the seeded agar plates and incubated in an inverted position, at 35°C for 16-18 hours and thereafter examined for clear zones of inhibition. Inhibition Zone Diameters (IZD) around each antibiotic disc were measured using a calibrated transparent ruler and recorded in millimetres. A standardized table was used to determine if each bacterium was ‘Resistant’, ‘Intermediate’ or ‘Sensitive’. [14] For the purpose of analysis, Isolates with intermediate or resistant results were merged as resistant. [15]
Quality control of culture and susceptibility testing was achieved using American Type Culture Collection (ATCC) standard reference strains Staphylococcus aureus (ATCC-25923), Escherichia coli (ATCC-25922), and Pseudomonas aeruginosa (ATCC 25853). Negative control was by a random selection of uninoculated culture media and incubation overnight for evidence of growth.
Data Analysis
Data were analysed with SPSS version 20(IBM Corporation, Armonk, NY, USA. Proportions of bacterial isolates and antibiotic sensitivities and resistances were presented as frequency tables. Prevalence rates of bacteria isolates were calculated as the frequency of identification of the bacterial species divided by the total number of all the bacteria species identified. Resistance rates were calculated for each antibiotic and each bacterial isolated by dividing the number of resistant isolates by the total number of isolates. [16] The overall resistance rates of each antibiotic were calculated as the number of bacteria resistant to antibiotic over the total number of bacteria isolates tested. [17] Chi-square test of association was used to compare the proportion of bacterial isolates with patients’ age and sex, with the level of significance p set as <0.05. Multiple Antibiotic Resistance (MAR) index was calculated for each isolate as the number of antibiotics to which the isolate is resistant/ Total number of antibiotics against which isolate was tested. [18]
Results
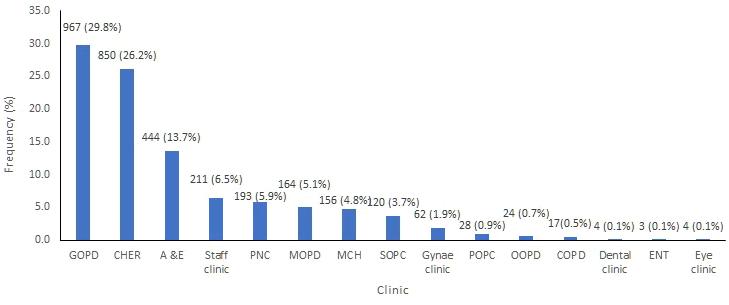
A total of 3,247 patient specimens from the out-patient clinics met the eligibility criteria. The samples were from females 1,581(48.7%) and from males 1,666(51.3%) The <9 years age group made up the highest proportion of patients accounting for 43.0%. Urine was the predominant specimen submitted,2058 (59.3%); followed by blood, 366 (19.6%). Out of 3,247 samples, 967 (42.5%) from the General Out-Patient Clinic (GOPD), 850 (52.3%) from the Children’s Emergency Room (CHER), 444 (13.7%) from the Accident and Emergency 164 (5.1%) from the Medical Out-Patient Clinic (MOPD). Other clinics from where samples were collected are shown in Figure 1.
Figure 1: Clinic name. GOPD: General Outpatient Clinic, CHER: Children’s Emergency Room, A & E: Accident and Emergency, PNC: Post-Natal Clinic, MOPD: Medical Out-Patient Clinic, MCH: Maternal and Child Health Unit, SOPC: Surgical Out-Patient Clinic, Gynae Clinic: Gynaecology Clinic, POPC: Paediatric Out-Patient Clinic, OOPD: Orthopedic Out-Patient Clinic, COPD: Consultants Out-Patient Clinic, ENT: Ear Nose and Throat Clinic.
Nine hundred and ninety-four (30.6%) samples showed significant microbial growth while 10 (0.3%) showed mixed growth and were excluded from further analysis. The remaining samples, 2,243(69.1%), either had no growth or insignificant growth. Four hundred and thirty-six (43.9%) isolates were Gram-positive, while 558(56.1%) isolates were Gram-negative. Urine yielded the most isolates 337(33.9%), followed by wound swab, 149 (15.0%), throat swab 120(12.1%) and sputum 115 (11.6%) [Table 1]. Most common bacterial species isolated was Escherichia coli 286 (28.8%), followed by Staphylococcus aureus 239 (24.0%) and Streptococcus pneumoniae 188 (18.9%), Others included Citrobacter species 50 (5.03%), Enterobacter species 62 (6.2%), Klebsiella species 53 (5.3%), Moraxella species 6 (0.6%), Proteus vulgaris 32 (3.2%), Pseudomonas aeruginosa 67 (6.7%), Serratiamarcella 1 (0.1%), Providencia species 1 (0.1%), Coagulase-negative staphylococcus species 6 (0.6%) and Xanthomonas species 1 (0.1%). Significantly more blood samples and wound swabs were received from males compared to females (p=0.02 and p=0.04 respectively), and urine from females compared to males (p=0.03). There was no significant association between sample type and sex for other samples. Isolates of Escherichia coli were significantly more predominant in samples from females compared to males (p=0.03) and Pseudomonas species in males compared to females (p<0.01) [Table 2].
| Variable | Frequency | Percentage |
|---|---|---|
| Type of specimen | ||
| Urine | 337 | 33.9 |
| Sputum | 115 | 11.6 |
| throat swab | 120 | 12.1 |
| Wound swab | 149 | 15 |
| Ear swab | 57 | 5.7 |
| Blood | 55 | 5.5 |
| Eye swab | 17 | 1.7 |
| ECS | 35 | 3.5 |
| HVS | 31 | 3.1 |
| Aspirate | 17 | 1.7 |
| Seminal fluid | 5 | 0.5 |
| Stool | 50 | 5 |
| Urethral swab | 5 | 0.5 |
| CSF | 1 | 0.1 |
| Sex distribution | ||
| Male | 436 | 43.9 |
| Female | 554 | 56.1 |
| Age group (years) | ||
| <9 | 517 | 52 |
| Oct-19 | 80 | 8.1 |
| 20-29 | 96 | 9.7 |
| 30-39 | 113 | 11.4 |
| 40-49 | 90 | 9.1 |
| >50 | 98 | 9.6 |
Table 1: Distribution of clinical Specimens (n=994).
| Variable | Female | Male | χ2 | P-value |
|---|---|---|---|---|
| Type of specimen | ||||
| Blood | 23 | 32 | 5.89 | 0.02* |
| CSF | 1 | - | Not applicable | |
| Ear swab | 30 | 27 | 0.6 | 0.44 |
| ECS | 35 | - | Not applicable | |
| Eye swab | 7 | 10 | 1.9 | 0.17 |
| HVS | 31 | - | Not applicable | |
| Pus aspirate | 9 | 8 | 0.15 | 0.7 |
| Seminal fluid | - | 5 | Not applicable | |
| Sputum | 70 | 45 | 0.59 | 0.44 |
| Stool | 22 | 28 | 3.95 | 0.05 |
| Throat swab | 61 | 59 | 2.52 | 0.11 |
| Urethral swab | - | 5 | Not applicable | |
| Urine | 209 | 128 | 4.18 | 0.04* |
| Wound swab | 74 | 75 | 4.46 | 0.04* |
| Type of Isolate | ||||
| Escherichia coli | 179 | 107 | 4.18 | 0.04* |
| Serratiaspecies | 1 | - | Not applicable | |
| Citrobacter species | 31 | 19 | 0.43 | 0.51 |
| Providencia species | 1 | Not applicable | ||
| Enterobacter species | 32 | 30 | 0.95 | 0.33 |
| Klebsiella species | 35 | 18 | 1.65 | 0.2 |
| Proteus vulgaris | 14 | 18 | 2.58 | 0.11 |
| Pseudomonas aeruginosa | 24 | 43 | 13.88 | <0.01* |
| Xanthomonas species | 1 | - | Not applicable | |
| Moraxella species | 4 | 2 | 0.21 | 0.65 |
| Coagulase-negative Staphylococcus | 4 | 2 | 0.21 | 0.65 |
| Streptococcus pyogenes | - | 2 | Not applicable | |
| Staphylococcus aureus | 143 | 96 | 0.67 | 0.41 |
| Streptococcus pneumoniae | 104 | 84 | 0.47 | 0.49 |
| *: Significant, NA: Not Applicable, CSF: Cerebrospinal Fluid, ECS: Endocervical Swab, HVS: High Vaginal Swab. | ||||
Table 2: Bivariate analysis of specimen type and Isolates by sex of patient.
Escherichia coli were the most frequently isolated pathogen accounting for 28.8% (286/994) of all isolates, and 51.3% of gram-negative pathogens (286/558). Staphylococcus aureus, 239 (54.8%) was the predominant gram-positive isolate. Staphylococcus aureus was the predominant isolate from blood 35 (62.5%), ear swab 24 (42.1%), endocervical swab 20 (57.1%), High Vaginal Swab (HVS) 15 (48.4%), pus aspirate 9 (52.9%), Seminal fluid 4 (80.0%), urethral swab 4 (80.0) and wound swab 64 (42.7%). Escherichia coli was the predominant isolate in urine 192 (57.0%) and stool 41 (82.0%). Predominant isolate from throat swab and sputum was Streptococcus pneumoniae, 98 (81.7%) and 72 (62.6%) respectively. Cerebrospinal fluid yielded Enterobacter species, 1 (100.0%) [Table 3].
| Isolates | Blood | CSF | Ear swab | ESC | Eye swab | HVS | pus | Seminal fluid | Sputum | Stool | Throat swab | Urethral swab | Urine | Wound swab |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gram-negative | ||||||||||||||
| Citrobacter species | 1 (1.7) | 1(2.6) | 1 (5.0) | 1 (3.2) | 3 (2.6) | 2 (4.0) | 2 (1.7) | 30 (8.9) | 9 (6.0) | |||||
| Escherichia coli | 5 (9.0) | 2 (3.5) | 6 (17.1) | 1 (5.9) | 9 (29.0) | 4 (23.2) | 1 (20.0) | 3 (2.6) | 41 (82.0) | 2 (1.7) | 192 (57.0) | 20 (13.4) | ||
| Enterobacter species | 1 (100.0) | 1 (1.7) | 2 (5.7) | 1 (5.9) | 2 (6.5) | 3 (17.6) | 7 (6.1) | 4 (8.0) | 3 (2.5) | 1 (20.0) | 24 (7.1) | 13 (8.7 | ||
| Klebsiella species | 4 (7.2) | 3 (5.7) | 17 (14.8) | 6 (5.0) | 19 (5.6) | 4 (2.7) | ||||||||
| Proteus species | 4 (7.0) | 1 (5.9) | 2 (1.7 | 2 (4.0) | 10 (3.0) | 13 (8.7) | ||||||||
| Providencia species | 1 (0.3) | |||||||||||||
| Pseudomonas aeruginosa | 1 (1.8) | 23 (40.4) | 1 (2.6) | 1 (2.0) | 1 (0.8) | 19 (5.6) | 21 (14.1) | |||||||
| Serratia species | 1 (0.7) | |||||||||||||
| Moraxellaspecies | 1 (1.8) | 1 (2.6) | 1 (0.9) | 1 (0.3) | 2 (1.3) | |||||||||
| Xanthomonas species | 1 (0.3) | |||||||||||||
| Gram-positive | ||||||||||||||
| Staphylococcus aureus | 35 (62.5) | 24 (42.1) | 20 (57.1) | 11 (64.7) | 15 (48.4) | 9 (52.9) | 4 (80.0) | 9 (7.8) | 8 (6.7) | 4 (80.0) | 36 (10.7) | 64 (43.0) | ||
| Coagulase-NegativeStaphylococcus species | 1 (1.8) | 1 (2.6) | 1 (3.2) | 1 (0.9) | 2 (0.6) | |||||||||
| Streptococcus pyogenes | 1 (5.9) | 1 (0.7) | ||||||||||||
| Streptococcus pneumoniae | 7 (12.7) | 2 (3.5) | 3 (8.6) | 2 (11.8) | 72 (62.6) | 98(81.7) | 1 (0.3) | 1 (0.7) | ||||||
| Grand Total | 55 (100.0) | 1 (100.0) | 57 (100.0) | 35 (100.0) | 17 (100.0) | 31 (100.0) |
17 (100.0) | 5 (100.0) | 115 (100.0) |
50 (100.0) |
120 (100.0) |
5 (100.0) | 337 | 149 (100.0) |
| CSF: Cerebrospinal Fluid, HVS: High Vaginal Swab | ||||||||||||||
Table 3: Distribution of bacterial isolates from clinical specimens (n =994).
Gram-positive pathogens generally showed high resistant rates to Cotrimoxazole, Tetracycline, Cloxacillin, Erythromycin (93.1%, 86.4%, 72.5%, and 68.1% respectively), and least resistance to Meropenem (0.0%), Retapamulin (0.0%), Azithromycin (0.0%), Cefixime (28.0%), Ceftazidime (35.8%), Ceftriaxone (24.5%), and Chloramphenicol (30.6%). All isolates had a MAR >0.2 [Table 4].
| Antibiotic | Staphylococcus aureus | Streptococcus pneumoniae | Coagulase-negative Staphylococcus | Total | ||||
|---|---|---|---|---|---|---|---|---|
| #T | R (%) | #T | R (%) | #T | R (%) | #T | R (%) | |
| Tetracycline | 23 | 18 (78.3) | 65 | 64 (98.5) | NT | 88 | 76 (86.4) | |
| Gentamicin | 225 | 82 (18.9) | 160 | 96 (60.0) | 6 | 3 (50.0) | 391 | 181 (46.3) |
| Cefuroxime | 173 | 56 (32.4) | 83 | 53 (63.9) | 5 | 3(60.0) | 261 | 112 (42.9) |
| Augmentin | 223 | 74 (33.2) | 176 | 89 (50.6) | 6 | 3 (50.0) | 405 | 166 (41.0) |
| Cloxacillin | 166 | 92 (37.3) | 115 | 111 (96.5) | 3 | 3 (100.0) | 284 | 206 (72.5) |
| Cefixime | 13 | 7 (53.8) | 12 | 0 (0.0) | NT | 25 | 7 (28.0) | |
| Ceftazidime | 205 | 72 (35.1) | 127 | 47 (37.0) | 6 | 2 (33.3) | 338 | 121 (35.8) |
| Ceftriaxone | 168 | 41 (24.4) | 117 | 29 (24.8) | 5 | 1 (20.0) | 290 | 71 (24.5) |
| Cotrimoxazole | 29 | 23 (79.3) | 73 | 72 (98.6) | NT | 102 | 95 (93.1) | |
| Amoxycillin | 7 | 3 (42.9) | 19 | 12 (63.2) | NT | 26 | 15 (57.7) | |
| Ofloxacin | 212 | 112 (52.8) | 123 | 50 (40.7) | 7 | 6 (85.7) | 342 | 168 (49.1) |
| Chloramphenicol | 16 | 7 (43.8) | 33 | 8 (24.2) | NT | 49 | 15 (30.6) | |
| Erythromycin | 183 | 100 (54.6) | 142 | 120 (84.5) | 4 | 4 (100.0) | 329 | 224 (68.1) |
| Streptomycin | 12 | 2 (16.7) | 35 | 27(77.1) | NT | 47 | 29 (61.7) | |
| Meropenem | 7 | 0 (0.0) | 10 | 0 (0.0) | NT | 17 | 0 (0.0) | |
| Clindamycin | 1 | 1 (100.0) | NT | NT | 1 | 1 (100) | ||
| Retapamulin | 1 | 0 (0.0) | 4 | 0 (0.0) | NT | 5 | 0 (0.0) | |
| Azithromycin | NT | 3 | 0 (0.0) | NT | 3 | 0 (0.0) | ||
| MAR index | 0.88 | 0.76 | 1 | |||||
| #: T number of isolates tested against each antimicrobial agent, R%: Percentage of isolates resistant to the antimicrobial agent, NT: Not Tested | ||||||||
Table 4: Antibiotic resistance profile of gram-positive bacteria isolates.
The Gram-negative isolates showed high rates of resistance to Erythromycin (95.6%), Amoxycillin (100.0%), Tetracycline (88.3%), Cloxacillin (96.9%), Amoxicillin–clavulanic acid (96.9%) and Cotrimoxazole (93.2%). The lowest resistances were shown against Nitrofurantoin (23.2%), Cefixime (17.5%) and Meropenem (0.0%). All isolates had MAR index >0.2 [Table 5].
| Antibiotic | Escherichia coli | Enterobacter species | Klebsiella species | Pseudomonas aeruginosa | Proteus species | Citrobacter species | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #T | R (%) | #T | R(%) | #T | R (%) | #T | R (%) | #T | R (%) | #T | R (%) | # T | R (%) | |
| Tetracycline | 36 | 33 (91.7) | 5 | 3 (60.0) | 12 | 10(83.3) | 3 | 3(100.0) | 2 | 2(100.0) | 2 | 2 (100.0) | 60 | 53 (88.3) |
| Gentamicin | 224 | 106(47.3) | 59 | 31 (52.5) | 51 | 16(31.3) | 63 | 37 (58.7) | 28 | 9 (32.1) | 46 | 27 (58.7) | 471 | 226 (48.0) |
| Cefuroxime | 163 | 110(67.5) | 46 | 38 (82.6) | 29 | 20 (68.9) | 48 | 46 (95.8) | 24 | 14 (58.3) | 32 | 20 (62.5) | 342 | 248(72.5) |
| Amoxicillin–clavulanic acid | 237 | 207(87.3) | 55 | 49(89.0) | 47 | 32(68.1) | 65 | 58(89.2) | 28 | 20 (71.4) | 46 | 43(93.5) | 478 | 409 (85.6) |
| Cloxacillin | 68 | 65 (95.6) | NT | NT | 26 | 26 (100.0) | NT | NT | 17 | 17 (100.0) | 18 | 17 (94.4) | 129 | 125 (96.9) |
| Cefixime | 52 | 9 (17.3) | 13 | 1(7.7) | 12 | 2 (16.7) | 9 | 2 (22.2) | 3 | 0 (0.0) | 8 | 3 (37.5) | 97 | 17 (17.5) |
| Ceftazidime | 204 | 95 (46.6) | 47 | 31 (65.9) | 43 | 19 (44.2) | 55 | 26 (47.3) | 27 | 6 (22.2) | 45 | 25 (55.6) | 421 | 202 (48.0) |
| Ceftriaxone | 116 | 48 (41.4) | 38 | 30 (78.9) | 30 | 8 (26.7) | 42 | 21 (50.0) | 22 | 4 (18.2) | 20 | 19 (95.0) | 268 | 130 (48.5) |
| Cotrimoxazole | 42 | 39(92.9) | NT | NT | 9 | 9(100.0) | 5 | 4 (80.0) | 2 | 2(100.0) | 1 | 1(100.0) | 59 | 55 (93.2) |
| Amoxycillin | 37 | 37 (100.0) | 3 | 3(100.0) | 5 | 5 (100.0) | 2 | 2 (100.0) | 2 | 2 (100.0) | 3 | 3 (100.0) | 52 | 52(100.0) |
| Ofloxacin | 232 | 114 (49.1) | 56 | 19 (33.9) | 45 | 13 (28.9) | 64 | 41 (64.1) | 28 | 11 (39.3) | 47 | 22 (46.8) | 472 | 220 (46.6) |
| Nitrofurantoin | 162 | 14 (8.6) | 18 | 5(27.8) | 16 | 8 (50.0) | 20 | 18(900.0) | 11 | 6 (54.5) | 23 | 7 (30.4) | 250 | 58 (23.2) |
| Meropenem | 20 | 0 (0.0) | 4 | 0 (0.0) | 6 | 0 (0.0) | NT | NT | 4 | 0 (0.0) | 1 | 0 (0.0) | 35 | 0 (0.0) |
| Ciprofloxacin | 47 | 37 (78.7) | NT | NT | 6 | 5 (83.3) | 10 | 9 (90.0) | NT | NT | 21 | 8 (38.1) | 84 | 59 (70.2) |
| Erythromycin | 75 | 71 (94.7) | 30 | 29 (96.7) | 26 | 26 (100.0) | 39 | 36 (92.3) | 20 | 19 (95.0) | 14 | 14 (100.0) | 204 | 195(95.6) |
| Nalidixic acid | 31 | 22(70.9) | 3 | 3 (100.0) | 3 | 3(100) | 2 | 2 (100) | NT | NT | 1 | 0 (0.0) | 40 | 30(75.0) |
| MAR index | 0.93 | 0.92 | 0.93 | 1 | 0.86 | 0.88 | ||||||||
Table 5: Antibiotic response to gram-negative organisms.
The resistance profiles of the isolates showed that all the isolates were resistant to at least one or more antimicrobial agents and a majority (75%) of the isolates were resistant to more than 3 antimicrobial agents.
Discussion
In the study, the majority of the specimens had no bacterial growth possibly because the patients may have taken antibiotics before coming to the clinic, as the practice of self-medication is high in country. [19] The higher proportion of isolates from females tallies with findings from other studies. [20,21]
In this study, Gram-positive pathogens were the predominant isolates, unlike other studies where Gram-negative pathogens dominated. [6,9,22,23] The high prevalence of Streptococcus pneumoniae in respiratory specimen has been similarly reported in other studies [24,25], at variance with a study in India where Klebsiella pneumoniae was the predominant isolate from respiratory tract specimens. [22] Streptococcus pneumoniae is responsible for 80% of community-acquired pneumonia across all age groups. [25,26] Streptococcus pneumoniae was found to have high rates of resistance to readily available first-line antibiotics and low rates of resistance to cephalosporins, carbapenems, Ofloxacin and Chloramphenicol. This finding corroborates a report from the Nigeria Centre for Disease Control. [27] A high rate of resistance to streptomycin is corroborated by other studies. [9] Contrary to this, Beyene et al. in their study in Ethiopia reported Streptococcus pneumoniae isolates with high resistance rates to oxacillin and low resistance to common firstline antibiotics. [24] High rates of resistance to carbapenems and quinolones have also been documented. [6]
The most common pathogen isolated from blood specimen was Staphylococcus aureus, in agreement with other studies. [9,23] The higher prevalence in the younger age group has similarly been documented. [28] Staphylococcus aureus showed high resistance to Amoxycillin, tetracycline, cotrimoxazole and low resistance to gentamycin, meropenem and Amoxycillin-clavulanic acid. Similar results have been reported. [23,24]
The high prevalence of E. coli isolates from urine specimen has been reported in other studies [29–31], and contrary to studies where Staphylococcus aureus [32] and Klebsiella spp [6] were the dominant uropathogens. E. coli was significantly isolated more from females than males, as similarly reported. [23,29,31] E. coli was found to be highly resistant to Tetracycline, Cotrimoxazole, Amoxycillin and Erythromycin and sensitive to Nitrofurantoin, gentamicin, Amoxycillin-clavulanic acid and the extended-spectrum Cephalosporins, in tandem with findings from other studies. [17,24,27,33,34] Nitrofurantoin, Gentamycin and Cephalosporins are indeed recommended for the empirical treatment of uncomplicated urinary tract infections and are available as oral preparations. [35] On the other hand, high resistance to Nitrofurantoin was observed in a study carried out in Cameroon. [30] Klebsiella species, the second common uropathogen in this study, showed high resistance to nalidixic acid and tetracycline and sensitivity to the cephalosporins. In contrast, high levels of resistance to cephalosporins have been documented in some studies. [6,10] The isolation of E. coli and Klebsiella species as primary pathogens responsible for urinary tract infection in this study agrees with other studies. [36] The frequency of isolation of other gram-negative pathogens (Citrobacter species, Proteus species, Pseudomonas aeruginosa, Serratiamarcella, Providencia species and Xanthomonas species) was low, consistent with other studies [32] and susceptibility ranged from highly sensitive to cephalosporins to resistant to common first-line antibiotics in tandem with some studies [34], and contrary to another study where resistance to cephalosporins was high. [6]
The finding in this study that gram-positive and negative bacterial pathogens are generally resistant to common inexpensive antibiotics is a reflection of the damage caused by inappropriate prescription practices including over-prescription and underprescription, misuse by the public fuelled by the availability of these cheap antibiotics over the counter and the sale of substandard antibiotics. This is in addition to the selection pressures that cause mutations and the spread of resistant strains in the community. Of note is the rising resistance to cefuroxime, as has been documented. [37]
Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species, have been given the acronym ESKAPE and as they tend to be multidrug-resistant and capable of “escaping” the biocidal action of antimicrobial agents. [38] They pose a threat in healthcare settings particularly among patients on invasive devices such as ventilators and blood catheters causing severe and often deadly bloodstream infections and pneumonia. Bacteria within the group have been demonstrated to be resistant to many antibiotics, including carbapenems and 3rd generation cephalosporins. [12] In this study, 4 of the ESKAPE pathogens were identified from the clinical samples, together making up 24.2% of the total isolates in the study. They included Staphylococcus aureus (15.9%), Klebsiella pneumoniae (6.2%), Pseudomonas aeruginosa (1.4%), and Enterobacter species (0.7%).
Conclusion
Gram-positive bacteria predominated among the outpatient’s samples tested. Gram-positive bacteria showed high resistance rates to Cotrimoxazole, Erythromycin, Cloxacillin, tetracycline, Amoxycillin-Clavulanic acid and Amoxycillin. Gram-negative bacteria showed high resistance to Tetracycline, Cotrimoxazole and Cloxacillin. The high rate of resistance to cefuroxime observed may be due to its availability over the counter, oral formation, poor dosing and poor compliance among the outpatient population.
Prescribers are left with a limited range of routinely used antibiotics to choose from as well as the increasing risk of resistance developing in the more expensive newer generation antibiotics. Hospital pharmacies should be stocked to reflect the current patterns of resistance to available antibiotics. Treatment guidelines should reflect the antibiotic resistance pattern to community-acquired infections. Interventions to reduce resistance including restrictions on over-the-counter sale of antibiotics should be strengthened. First-line antibacterial drugs showing marked reduction of efficacy should be withdrawn and reintroduced after a few decades (Antimicrobial recycling).
Ethical Consideration
Ethical approval was obtained from the Ethics committee of Irrua Specialist Teaching Hospital. Patients names were not entered into the data extraction sheet, and all other required demographic information as well as information on bacterial isolates were kept confidential by the researchers.
Acknowledgements
The authors are grateful to Prof Danny Asogun and PANDORA for funding the data collection and publication, the Resident Doctors, Medical Laboratory Scientists and laboratory technicians who took part in the laboratory analysis of the clinical samples, and the ISTH staff who assisted with data retrieval.
Funding
Funding for data collection, analysis, and article processing charge was provided as part of the PANDORA–ID-NET EDCTP Reg/Grant RIA2016E-1609. The grant is funded by the European and Developing Countries Clinical Trials Partnership (EDCTP2) program which is supported under Horizon 2020 the European Union’s Framework Program for Research and Innovation. The views and opinions of the author herein do not necessarily state or reflect those of EDCTP.
REFERENCES
- Nagel TE, Chan BK, De Vos D, El-Shibiny A, Kang’ethe EK, Makumi A, et al. The developing world urgently needs phages to combat pathogenic bacteria. Front Microbiol. 2016;7:882.
- World Health Organisation. Antimicrobial resistance global report on surveillance. Geneva. 2014.
- Hawkey PM, Warren RE, Livermore DM, McNulty CAM, Enoch DA, Otter JA, et al. Treatment of infections caused by multidrug-resistant gram-negative bacteria: Report of the British society for antimicrobial chemotherapy/healthcare infection society/British infection association joint working party. J Antimicrob Chemother. 2018;73:iii2-78.
- Lewis K. Recover the lost art of drug discovery. Nature. 2012;485:439-440.
- Dye C. After 2015: Infectious diseases in a new era of health and development. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130426.
- Shanthi DCM, kumar AS, Bharathi N. Study of prevalence and antimicrobial susceptibility pattern of bacterial isolates in a tertiary care hospital. Int J Pharm Pharm Sci. 2015;7:185-190.
- Aboderin OA, Abdu A-R, Odetoyin BW, Lamikanra A. Antimicrobial resistance in Escherichia coli strains from urinary tract infections. J Natl Med Assoc. 2009;101:1268-1273.
- Eucharia N, Chioma O, Eunice A. Oprd Genes detected in pseudomonas aeruginosa isolates from a teaching hospital but lost in a carbapenem-resistant strain. J Adv Med Med Res. 2019;29:1-8.
- Okesola A, Oni A. Antimicrobial resistance among common bacterial pathogens in South Western Nigeria. Am J Agric Environ Sci. 2009;5:327-330.
- Kumburu HH, Sonda T, Mmbaga BT, Alifrangis M, Lund O, Kibiki G, et al. Patterns of infections, aetiological agents and antimicrobial resistance at a tertiary care hospital in northern Tanzania. Trop Med Int Heal. 2017;22:454-464.
- Leber A. Clinical microbiology procedures handbook. (4th ed) American Society of Microbiology; 2016.
- Trojan R, Razdan L, Singh N. Antibiotic susceptibility patterns of bacterial isolates from pus samples in a tertiary care hospital of Punjab, India. Int J Microbiol. 2016;2016.
- Tille P. Bailey & Scott’s diagnostic microbiology. (13th ed) Missouri: Mosby Elsevier. 2013.
- Institute SC and L. Performance standards for antimicrobial susceptibility testing. (29th ed) Pennsylvania, 2016.
- Turnidge J, Paterson DL. Setting and revising antibacterial susceptibility breakpoints. Clin Microbiol Rev. 2007;20:391-408.
- Gufe C, Hodobo TC, Mbonjani B, Majonga O, Marumure J, Musari S, et al. Antimicrobial profiling of bacteria isolated from fish sold at informal market in Mufakose, Zimbabwe. Int J Microbiol. 2019;2019.
- Hove R-J ten, Tesfaye M, Hove WF ten, Nigussie M. Profiling of antibiotic resistance of bacterial species recovered from routine clinical isolates in Ethiopia. Ann Clin Microbiol Antimicrob. 2017;16:46.
- Krumperman PH. Multiple antibiotic resistance indexing of escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol. 1983;46:165-170.
- Abdulraheem IS, Adegboye A, Fatiregun AA, Syed, Rizvi AA. Self-medication with antibiotics: Empirical evidence from a Nigerian rural population. Br J Pharm Res. 2016;11:1-13.
- Magliano E, Grazioli V, Deflorio L, Leuci AI, Mattina R, Romano P, et al. Gender and age-dependent etiology of community-acquired urinary tract infections. Sci World J. 2012;2012.
- Abebe M, Tadesse S, Meseret G, Derbie A. Type of bacterial isolates and antimicrobial resistance profile from different clinical samples at a Referral Hospital, Northwest Ethiopia: Five years data analysis. BMC Res Notes. 2019;12:568.
- Santosh KA, Hatkar SS, Siddique S, Deshmukh A, Afreen U, Mariya S. Bacteriological profile and antimicrobial sensitivity pattern of clinical isolates from patients attending tertiary care hospital. Ann Pathol Lab Med. 2016;3:1-6.
- Mulu W, Abera B, Yimer M, Hailu T, Ayele H, Abate D. Bacterial agents and antibiotic resistance profiles of infections from different sites that occurred among patients at Debre Markos Referral Hospital, Ethiopia: A cross-sectional study. BMC Res Notes. 2017;10:254.
- Beyene G, Regasa B, Yilma D, Sewunet T. Antimicrobial susceptibility pattern of bacterial isolates from community-acquired pneumonia patients in Jimma University Specialized Hospital, Jimma, Ethiopia. Saudi J Heal Sci. 2015;4:59.
- Fiberesima F, Onwuchekwa AC. Community acquired pneumonia in Port Harcourt Rivers State of Nigeria. Cent Afr J Med. 2008;54:1-7.
- Biscevic-Tokic J, Tokic N, Musanovic A. Pneumonia as the most common lower respiratory tract infection. Med Arch. 2013;67:442-445.
- Nigeria Centre for Disease Control. Antimicrobial use and resistance in Nigeria situation analysis and recommendations. Abjua Nigeria: 2017.
- Nwankwo EO. Antibiotic sensitivity pattern of Staphylococcus aureus from clinical isolates in a tertiary health institution in Kano, Northwestern Nigeria. Pan Afr Med J. 2011;8.
- Bishop GH, Shehu F, Author C. Prevalence and antibiotic susceptibility patterns of bacterial etiologies of urinary tract infections among students attending Sick-Bay of Ahmadu Bello University, Nigeria. Edorium J Microbiol, 2016;2:7-12.
- Akoachere J-FTK, Yvonne S, Akum NH, Seraphine EN. Etiologic profile and antimicrobial susceptibility of community-acquired urinary tract infection in two Cameroonian towns. BMC Res Notes. 2012;5:219.
- Oluwafemi T, Akinbodewa A, Ogunleye A, Adejumo O. Urinary tract infections and antibiotic sensitivity pattern of uropathogens in a tertiary hospital in South West, Nigeria. Sahel Med J. 2018;21:18.
- Otajevwo FD, Eriagbor. Asymptomatic urinary tract infection occurrence among students of a private university in western delta, Nigeria. 2014.
- Kadri SM, Gash B, Rukhsana A. Antibiotic sensitivity and resistance profile of the micro-organisms responsible for urinary tract infection observed in Kashmir, India. J Sci Tech. 2010;11:61-72.
- Mukhtar AM, Saeed HA. Profile of antibiotic sensitivity and resistance of some pathogenic bacteria isolated from clinical specimens in Sudan. J Sc Tech. 2010;11:61-72.
- Ábrók G, Burián L. Comparative epidemiology and resistance trends of common urinary pathogens in a tertiary-care hospital: A 10-year surveillance study. Medicina. 2019;55:356.
- Behzadi P, Behzadi E, Yazdanbod H, Aghapour R, Cheshmeh AM, Omran SD. A survey on urinary tract infections associated with the three most common uropathogenic bacteria. Maedica (Buchar). 2010;5:111-115.
- Nsofor CA, Iroegbu CU. Antibiotic resistance profile of Escherichia coli isolated from five major geopolitical zones of Nigeria. J Bacteriol Res. 2013;5:29-34.
- Navidinia M. The clinical importance of emerging eskape pathogens in nosocomial infections. J Paramed Sci. 2016;7:43-57.




 The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.
The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.