Carfilzomib Triggers Endoplasmic Reticulum Stress By Up-Regulating the PERK-eIf2α Pathway in Uterine Corpus Endometrial Carcinoma Cells
2 School of Pharmacy of Xi'an Jiaotong University, Xi'an, 710061, China
3 Department of Pathogenic Biology and Immunology, Health Science Center of Xi'an Jiaotong University, Xi'an, 710061, China
Received: 22-Jun-2022, Manuscript No. AMHSR-22-69923; Editor assigned: 24-Jun-2022, Pre QC No. AMHSR-22-69923; Reviewed: 08-Aug-2022 QC No. AMHSR-22-69923; Revised: 27-Jan-2023, Manuscript No. AMHSR-22-69923; Published: 03-Feb-2023, DOI: 10.54608.annalsmedical.2023.83
Citation: Feng Z, et al. Carfilzomib Triggers Endoplasmic Reticulum Stress By Up-Regulating the PERK-eIf2α Pathway in Uterine Corpus Endometrial Carcinoma Cells. Ann Med Health Sci Res. 2023;13:431-438.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Uterine corpus endometrial carcinoma is one of the most common female pelvic malignancies. Carfilzomib, a second-generation Proteasome Inhibitor (PI), is currently being used for the treatment of multiple myelomas by inhibiting the pathways of IRE1-XBP1 and PERK-eIf2α. However, when used to treat solid tumors, carfilzomib activated the XBP1-IRE1 pathway without affecting PERK-eIf2α. This study aimed to elucidate the function of carfilzomib in UCEC cells and to uncover the influence of PIs on specific cancer cells. Here, two cell lines were treated with carfilzomib, and cell viability, and protein and gene expressions were evaluated. In addition, tumor-carrying BALB/c immunodeficint mice were treated with carfilzomib with or without UMI-77 supplementation, and tumor growth was analyzed. We found that Carfilzomib treatment increased apoptosis and the expression of ER stress signaling markers and pro-apoptotic proteins (e.g., Noxa and Puma) increased, whereas the expression of anti-apoptotic proteins (e.g., Bcl-2 and Bcl-xl) decreased. The expression of markers related to three different ER stress pathways were affected by carfilzomib treatment. In nude mice models, carfilzomib function was more pronounced with UMI-77 supplementation. To conclude, we have shown that Carfilzomib triggers ER stress in UCEC cells in vitro and leads to tumor necrosis in vivo.
Keywords
Carfilzomib; Endoplasmic reticulum stress; Uterine corpus endometrial carcinoma; Apoptosis
Introduction
Poly Cystic Uterine Corpus Endometrial Carcinoma (UCEC) is a global public health problem and is the most prevalent gynecological malignancy in developed countries. According to statistical analyses, the prognosis of UCEC is better than that of ovarian and cervical cancer because UCEC can be detected earlier due to the early appearance of symptoms (e.g. irregular vaginal bleeding). The 5 years survival rate of UCEC is up to 90% following surgery. However, 15–25% of the patients with advanced UCEC show unfavorable prognoses due to a lack of new adjuvant therapeutic modalities [1-3]. With increasing incidence in recent years, more studies are required to understand the underlying mechanisms of UCEC to aid the development of treatment strategies [4]. Even though UCEC can be diagnosed at an early stage, it often cooccurs with comorbidities, which raises the risk of traumatic surgery. Therefore, exploring new anti-tumor therapeutics with high specificity and sensitivity will be of clinical significance. The Endoplasmic Reticulum (ER) is central to protein synthesis and transportation and plays a role in maintaining a stable cellular microenvironment (ER homeostasis). The proteasome is responsible for the degradation of intracellular proteins [5-9]. A malfunction or the disruption of the proteasome can disrupt ER homeostasis and lead to the accumulation of misfiled and unfolded proteins. The ER stress signaling pathway can be activated by at least three signaling pathways, which ultimately leads to cell death under stress. Proteasome Inhibitors (PIs) are a series of drugs that block the action of the proteasome by selectively and irreversibly binding to chymotrypsin like sites in the proteolysis core. Since PIs can disrupt cell cycle regulation processes, they are used to induce programmed cell death and reduce the viability of cancer cells via classical and/or ER stress pathways. Studies have revealed the involvement of different signaling pathways in multiple myeloma, a kind of plasma malignant tumor and adrenocortical carcinoma, a kind of solid tumor, revealing the divergent behaviors of tumor cells under the pressure of ER stress induced by PIs. Carfilzomib, a PI, has been shown to significantly reduce cell viability and is currently being used as a pharmacotherapy for the treatment of multiple myelomas. We have previously shown that the treatment of UCEC cells with Carfilzomib led to the overexpression of Myeloid cell leukemia-1 (Mcl-1), an anti-apoptosis protein of the B-cell lymphoma-2 (Bcl-2) family, and led to cell cycle arrest. The response of UCEC cells to Carfilzomib was investigated to understand how UCEC cells differentiate compared to the cancers mentioned above [10-13]. Here, we explored the cytotoxic activity and potential mechanism of Carfilzomib in HEC-1-A and Ishikawa cell lines in BALB/c immunodeficient mice. In vitro and in vivo experiments revealed that Carfilzomib significantly induced cell apoptosis and the accumulation of Mcl-1 via ER stress signaling pathways. Moreover, the behavior of Carfilzomib in vivo indicated its potential chemotherapeutic function. Which was significantly enhanced in combination with UMI-77, a selective inhibitor of Mcl-1?
Materials and Methods
Drugs and antibodies
Carfilzomib, purchased from LC laboratories (Woburn, MA, USA), was reconstituted in Dimethyl Sulfoxide (DMSO) at a stock concentration of 10 mmol/L and stored at −20°. The stock was diluted in DMSO to the needed concentrations (0 nM (no Carfilzomib, DMSO only), 12.5 nM, 25 nM, 50 nM, 100 nM). The final concentration of DMSO was <0.01 % in the cell culture medium. The cells were cultured in Dulbecco's modified eagle medium (DMEM; Sigma-Aldrich; Schnelldorf, Germany) supplemented with 10 % (w/v) heat inactivated fetal bovine serum (FBS; HyClone; Logan, UT, USA), 100 U/mL penicillin, and 50 g/mL streptomycin at 37°C in 5 % CO2. The primary antibodies (rabbit) used in this study included: anti-Bcl-2, anti-Mcl-1, anti-Bcl-x1, antibax, antibim, antipuma, anti-β-actin, antinoxa, anti-GRP78 (glucose-regulated protein 78), anti-CHOP (C/EBPhomologous protein), anti-PERK (protein kinase R-like ER kinase), anti-p-eIF2α (eukaryotic initiation factor 2), anti-p- PERK, anti-XBP1 (X-box binding protein 1), anti-ATF4 (activating transcription factor 4), and anti-ATF6. All the primary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The secondary HRP conjugated goat anti-rabbit antibodies were purchased from cell signaling technology (Boston, MA, USA).
Cell culture
The HEC-1-A (moderately differentiated endometriosis, more sensitive to Carfilzomib) and Ishikawa cell (welldifferentiated endometriosis, less sensitive to Carfilzomib) lines were obtained from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in DMEM (Sigma-Aldrich; Schnelldorf, Germany) supplemented with 10 % (w/v) heat inactivated FBS (HyClone; Logan, UT, USA), 100 U/mL penicillin, and 50 g/mL streptomycin at 37°C in 5 % CO2.
Flow cytometry
Apoptotic cells were evaluated using the annexing V–FITC apoptosis detection kit (BD Biosciences, NJ, and USA). All experimental steps were performed in accordance with the protocols of manufacturer. Briefly, 1 × 106 cells were washed twice with Phosphate-Buffered Saline (PBS) and stained with 100 μL of binding buffer containing 5 μL of 7-aminoactinomycin D and 5 μL of annexin V–FITC. The cells were incubated at 4°C for 15 min, suspended in 400 μL of binding buffer, and immediately analyzed using a Fluorescence Activated Cell Sorting (FACS) flow cytometer (BD Biosciences). Ten thousand events were recorded and analyzed [14].
Western blotting
All experimental steps were performed in accordance with the protocols of manufacturer. Briefly, cells were lysed with Radio Immuno Precipitation Assay (RIPA) buffer (Roche Applied Science; Mannheim, Germany) containing a protease inhibitor cocktail and phenylmethylsulfonyl fluoride. Protein concentrations were determined using the Bicin Choninic Acid (BCA) protein assay kit (Pierce; Rockford, IL, USA). Equal amounts of proteins were separated using 12% Sodium Dodecyl Sulfate- Polyacrylamide Gel Electrophoresis (SDS-PAGE) and electrophoretically transferred to a Polyvinylidene Fluoride (PVDF) membrane (Millipore Corporation; Bedford, MA, USA). The membranes were blocked for one hour at room temperature with 5% nonfat milk. Next, the membranes were incubated with primary antibodies against Bax, Mcl-1, Bcl-2, Bcl-x1, Bim, Puma, Noxa, GRP78, CHOP, PERK, p-eIF2α, p-PERK, XBP1, ATF4, and ATF6 overnight at 4°C. The membranes were washed four times and incubated with antirabbit IgG (1:1,000) conjugated to Herseradish Peroxidase (HRP) at 37°C for one hour. Blots were visualized using Enhanced Chemiluminescence (ECL) reagents (Millipore Corporation; Bedford, MA, USA).
Quantitative RT-PCR
All experimental steps were performed in accordance with the protocols of manufacturer. Briefly, RNA extraction was performed using the RNeasy kit (QIAGEN, Dusseldorf, Germany), and the isolated Total RNA was reversetranscribed by the Verso cDNA synthesis kit (Thermo Scientific, MA, USA), and qRT-PCRs were performed using the Maxima SYBR Green/ROX qPCR Master Mix (Fermatas) on an 7900 HT fast real time PCR system from thermo fisher scientific (Waltham, MA, USA) with genespecific primers [15-19]. The cycling conditions were as follows: two min at 50°C followed by ten min at 95°C; 40 cycles of 95°C for 15 sec and 60°C for one min. Cycle threshold (Ct) values were collected for β-actin and the genes of interest during the log phase of the cycle. Quantification of target gene expression was normalized against β-actin using the Ct method (Tables 1 and 2).
| Table 1: Primer sequences used for Qrt-PCR. | ||
|---|---|---|
| Gene | Forward sequences (5’-3’) | Reverse sequences (5’-3’) |
| ß-actin | ATCGTGCGTGACATTAAGGAGAAG | AGGAAGGAAGGCTGGAAGAGTG |
| Bcl-2 | TTCAACACAGACCCACCCAGAG | GCAGGATAGCAGCACAGGATTG |
| Bcl-xl | GCAGCCGAGAGCCGAAAGG | GGATGTGGTGGAGCAGAGAAGG |
| Mcl-1 | AAGAGGCTGGGATGGGTTTGTG | TTGGTGGTGGTGGTGGTTGG |
| Puma | CTGCCGCCCACCACCATC | TGAAGGAGCACCGAGAGGAGAG |
| Noxa | ACCGTGTGTAGTTGGCATCTCC | AGGTTCCTGAGCAGAAGAGTTTGG |
| GRP78 | AGGAGGAGGACAAGAAGGAGGAC | CAGGAGTGAAGGCGACATAGGAC |
| CHOP | TGCTTCTCTGGCTTGGCTGAC | CCGTTTCCTGGTTCTCCCTTGG |
| XBP1 | TTGCTGAAGAGGAGGCGGAAG | GGTCCAAGTTGTCCAGAATGC |
| Table 2: Inhibitory function of cfz/umi-77 in the tumor-carrying nude mice models (n=3 in each group, *p<0.05,**p<0.01). | ||||
|---|---|---|---|---|
| Groups | Initial body weight (g) | Final body weight (g) | Final tumor weight (g) | Inhibitory rate (%) |
| Control | 23.85 ± 1.31 | 22.68 ± 0.97 | 0.74 ± 0.41 | - |
| CFZ | 22.55 ± 2.09 | 21.00 ± 2.21 | 0.53 ± 0.28* | 28.37* |
| UMI-77 | 22.95 ± 1.17 | 21.55 ± 1.35 | 0.68 ± 0.16 | 8.1 |
| CFZ+UMI-77 | 20.53 ± 1.73 | 19.33 ± 2.22 | 0.36 ± 0.20** | 51.35** |
Cell viability assays
After standard cell cultivation for 24 h, the HEC-1-A and Ishikawa cell lines were seeded in a 96-well plate (8 × 103 cells per well) with graded doses of medicine for a further 24 h and subjected to standard MTT analysis. Briefly, 200 μl of solution (containing 0.5 mg/mL of MTT, Omega Biotech (Norcross, GA, USA)) was added at the indicated time points at 37°C. After cell culture for four h, cells per well were lysed in 150 μl of DMSO for 15 min at room temperature. The inhibitory rate (%) was evaluated by an FLUO star OPTIMA enzyme labeled instrument from the (BMG LABTECH, (Offenburg, Germany) at OD490.
Animals and drug treatments
Female BALB/c immunodeficient mice aged 4-6 weeks, weighing 15 g-18 g, were purchased from the Shanghai lab animal research center (Shanghai, China). The mice were kept in a clean animal room of the experimental animal center of the medical department of Xi'an Jiao Tong University and had free access to food and water. A nude mouse model was established by subcutaneously injecting 1 × 107 HEC-1-A cells/mL (0.2 ml). After about 1.5 weeks, the mice were divided into four groups according to treatment: control group (0.5 % Sodium salt of Carboxy Methyl Cellulose), Carfilzomib (2.5 mg/kg), UMI-77 (50 mg/kg), Carfilzomib and UMI-77 (Carfilzomib 2.5 mg/kg+UMI-77 50 mg/kg). The drugs were administered for 2 weeks, and tumor volumes were measured every other day [20].
Hematoxylin and eosin (H and E) staining
Autopsies were performed on euthanized mice to remove the tumor tissues. The tissues were temporarily stored in paraformaldehyde (PFA). The samples were placed in processing cassettes, dehydrated using a serial alcohol gradient (50%, 75%, 80%, 95%, and 100%) and xylene, and embedded in 62°C paraffin wax blocks. Next, 4 μm sections were dewaxed in xylene twice, rehydrated using decreasing concentrations of ethanol (100%, 95%, 80%, and 70%), and finally washed and immersed in distilled water. Next, they were stained with hematoxylin and washed with water. This was followed by differentiation and bluing and then staining with eosin and lastly washing with distilled water. Then the sections were dehydrated using a serial alcohol gradient (50%, 75%, 80%, 95 %, and 100%) and xylene. Finally, sections were observed under an optical microscope at 100X and 400X magnifications [21-23].
Statistical analysis
Data are presented as Mean ± Standard Error (SE). A one way Analysis of Variance (ANOVA) was used to compare the means of the different testing groups and to calculate the p-value. Significance was set at p<0.05. The Coefficient Indices (CIs) of Carfilzomib (nM) and UMI-77 (μM) were calculated using the computer programmer CompuSyn by entering the inhibitory rates of the different groups.
Results
Carfilzomib induces apoptosis in a dose-dependent manner and modulates the expression of bcl-2 family proteins
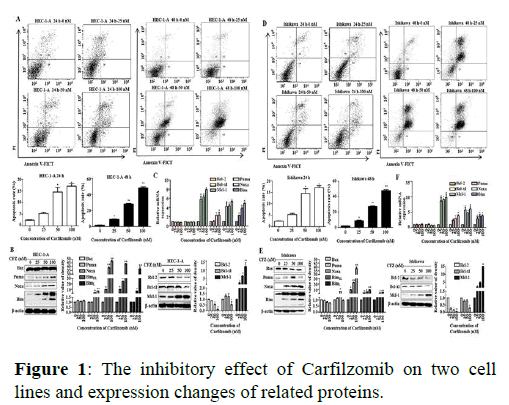
We have previously shown that Carfilzomib has dosedependent anti-proliferative effects on cell cycle arrest in G2/M cells by down regulating Cyclin D3 in UCECs. Here we performed annexing V–FITC and PI fluorescence staining to determine whether ER stress can induce cell apoptosis in cell lines following Carfilzomib treatment. Signals were barely detected in the control groups of HEC-1-A and Ishikawa cells (Figure 1).
Note: (A) Carfilzomib triggered dose-dependent apoptosis of HEC-1-A cells after treatment for 24 h and 48 h, the cells apoptosis was evaluated by flow cytometry and cells apoptosis rate chart were showed. FACS data shown are representative of three independent experiments. n=3 for all experiments. The 0 μM group meant the final concentration of drug (Carfilzomib) in the cell culture medium was 0 nM, only pure solvent (DMSO) was added. (B) Bcl-2 family anti-apoptotic and pro-apoptotic proteins in HEC-1-A cells after 24 h treatment with graded doses of Carfilzomib. It showed the anti-apoptotic proteins expression of Bcl-2 family in HEC-1-A by Western Blot. And the relative quantification of the protein in Bcl-2 family proteins was shown below (n=3).(C) The mRNA of Bcl-2 family members in HEC-1-A after 24 h treated by Carfilzomib. *P<0.05**, P<0.01. Carfilzomib triggered dose-dependent apoptosis of Ishikawa cells after treatment for 24 h and 48 h, the cells apoptosis was evaluated by flow cytometry and cells apoptosis rate chart were showed. (D) FACS data shown are representative of three independent experiments. n=3 for all experiments. The 0 μM group meant the final concentration of drug (Carfilzomib) in the cell culture medium was 0 nM, only pure solvent (DMSO) was added. (E) Bcl-2 family anti-apoptotic and pro-apoptotic proteins in Ishikawa cells after 24 h treatment with graded doses of Carfilzomib. It showed the anti-apoptotic proteins expression of Bcl-2 family in Ishikawa by Western Blot. And the relative quantification of the protein in Bcl-2 family proteins was shown below (n=3).(F) The mRNA of Bcl-2 family members in Ishikawa after 24 h treated by Carfilzomib. *P<0.05, **P<0.01.
Significant drug induced cell apoptosis was detected after 24 h and 48 h treatments with graded concentrations of Carfilzomib (25 nM, 50 nM, and 100 nM). To further explore the molecular mechanism of apoptosis induced by Carfilzomib, qRT-PCR was used to evaluate the expression of anti- and pro-apoptotic genes. Carfilzomib unregulated the expression of Mcl-1, Bim, Puma, and Noxa, but had no significant effect on the expression of Bcl-2 and Bcl-xl. The qRT-PCR results were supplemented with western blotting, to check protein accumulation following treatment. Carfilzomib significantly increased the expression of Mcl-1, Bim, Puma, and Noxa, but had no significant effect on the expression of Bax in two cell lines.
CarfilzomibdifferentiallyaffectsthedifferentER stresspathways
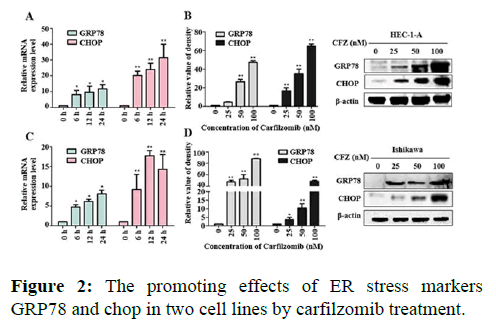
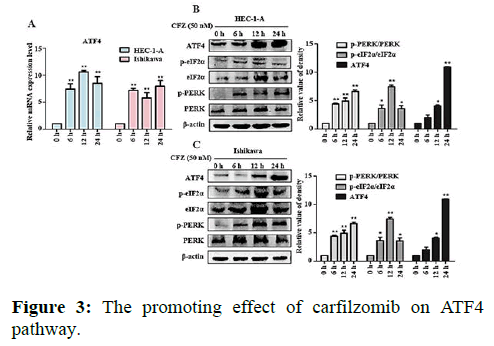
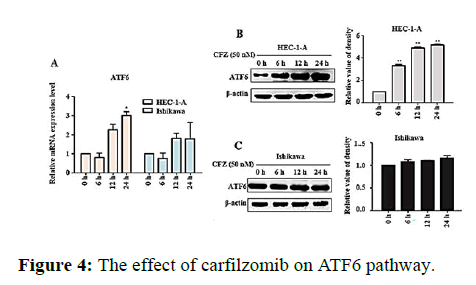
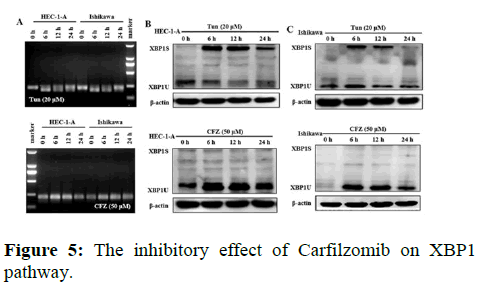
We next evaluated the effect of Carfilzomib on the expression of two ER stress markers, GRP78 and CHOP. Quantitative RT-PCR revealed that 50 nM of Carfilzomib significantly unregulated the expression of GRP78 and CHOP in a time dependent manner. Moreover, western blotting revealed an up regulation of GRP78 and CHOP in a dose dependent manner with 24 h of Carfilzomib treatment. Stimulation of the Unfolded Protein Response (UPR) by ER stress activates three key pathways that ultimately initiate the CHOP apoptotic pathway. These three pathways include the PERK, ATF6, and IRE-1α (inositol-requiring enzyme 1) pathways. We evaluated the expression of marker genes/proteins associated with these three pathways. Stimulation of the PERK pathway can lead to phosphorylation of eIF2α, which promotes the expression of ATF4, a key molecule in the PERK pathway. Quantitative RT-PCR showed that the expression of ATF4 was significantly unregulated following Carfilzomib treatment. At the protein level, Carfilzomib elevated the levels of eIF2α and PERK phosphorylation and induced the expression of ATF4. Regarding the ATF6 pathway, ATF6 expression was significantly unregulated in a time-dependent manner in HEC-1-A cells following Carfilzomib treatment but not in Ishikawa cells. At the protein level, ATF6 expression was increased in the HEC-1-A cells but not in the Ishikawa cells. Tunicamycin, often used as an inhibitor of ER N-linked glycosylation, can specifically induce cellular ER stress and activate the IRE1α-XBP1 pathway. Agarose gel electrophoresis showed that the expression of XBP1S was unchanged following Carfilzomib treatment. At the protein level, the precursor form of XBP1, XBP1U was elevated in the Carfilzomib-treated groups, but the activation form, XBP1S, was not (Figures 2-5).
Note: (A) Effects of mRNA of GRP78, CHOP in HEC-1-A after 24 h treated by Carfilzomib *P<0.05, **P<0.01. (B) Western Blot showed the protein expression situations of GRP78 and CHOP in HEC-1-A and the relative quantifications of the GRP78, CHOP proteins in HEC-1-A cells were evaluated (n=3). (C) Effects of mRNA of GRP78, CHOP in Ishikawa after 24 h treated by Carfilzomib *P<0.05, **P<0.01. (D) Western Blot showed the protein expression situations of GRP78 and CHOP in Ishikawa and the relative quantifications of the GRP78, CHOP proteins in Ishikawa cells were evaluated (n=3).
Note: (A) Effects of ATF4 mRNA in HEC-1-A and Ishikawa treated by Carfilzomib. (B) Effects of protein expression in upstream of ATF4 pathway in HEC-1-A by Carfilzomib treated. ATF4 Protein changed in HEC-1-A after Carfilzomib treated in different time and the relative quantification of the proteins in HEC-1-A cells (n=3). (C) Effects of protein expression in upstream of ATF4 pathway in Ishikawa by Carfilzomib treated. ATF4 Protein changed in Ishikawa after Carfilzomib treated in different time and the relative quantification of the proteins in Ishikawa cells (n=3) *P<0.05, **P<0.01.
Note: (A) The result of relative mRNA expression levels of ATF6 mRNA in HEC-1-A and Ishikawa changed with different doses of Carfilzomib. (B) The expression of protein ATF6 by Carfilzomib treated in different time in HEC-1-A shown in Western blot and the relative quantification of the proteins (n=3). (C) CThe expression of protein ATF6 by Carfilzomib treated in different time in Ishikawa shown in Western blot and the relative quantification of the proteins (n=3). *P<0.05, **P<0.01.
Note: (A) The result of agarose gel electrophoresis showed the effect of tunicamycin and Carfilzomib on XBP1 transcription in HEC-1-A and Ishikawa cells. (B) The expression of XBP1 (XBP1S and XBP1U) protein in HEC-1-A by tunicamycin and Carfilzomib treatment shown in Western blot and the relative quantification of the proteins in HEC-1-A cells (n=3). (C) The expression of XBP1 (XBP1S and XBP1U) protein in HEC-1-A by tunicamycin and Carfilzomib treatment shown in Western blot and the relative quantification of the proteins in Ishikawa cells (n=3) *P<0.05,**P<0.01.
Carfilzomib inhibits the proliferation of UCEC cells in vivo , especially in combination with umi-77
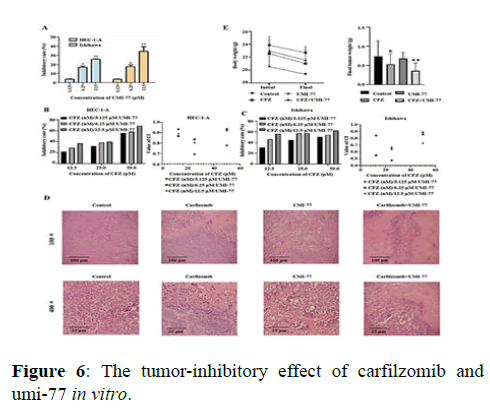
The MTT analysis showed that Carfilzomib inhibited the growth of UCEC cells, with the IC50 (half maximal inhibitory concentration) values of 24 h treated HEC-1-A and Ishikawa cells to be 7.18 × 10-8 mol/L and 1.20 × 10-7 mol/L, respectively. Regarding its application in chemotherapy, the graded concentrations of Carfilzomib were determined to be 12.5 nmol/L, 25 nmol/L, and 50 nmol/L to achieve an optimal concentration in combination with UMI-77. The Mcl-1 selective inhibitor, UMI-77, was able to inhibit the proliferation of HEC-1-A and Ishikawa cells when applied in the range of 3.725 to 50 μmol/L. Statistically, the IC50 values of UMI-77 for HEC-1-A and Ishikawa cells were 3.07 × 10-5 mol/L and 2.863 × 10-5 mol/L, respectively. Thus, the following concentrations of UMI-77 were used in combination with Carfilzomib for further experiments: 3.125 μmol/L, 6.25 μmol/L, and 12.5 μmol/L. The MTT analysis showed that inhibitory rates of the UMI-77 treated group were increased in a dose-dependent manner in vitro. The inhibitory rate was evaluated via MTT analysis, and the Coefficient Index (CIs) of the drugs that were administrated in vitro was calculated by CompuSyn. Results showed that all the CIs of the combination-treated gradients were lower than 1.0. Regarding the in vivo experiments, the tumor inhibitory rate was 28.37 % in the Carfilzomib only group, 8.10 % in the UMI-77 only group, and 51.35 % in the combination group (UMI-77+Carfilzomib). Moreover, H and E staining of tumor tissues showed evident atypia of UCECs with large nuclei and red stained cytoplasm in the treated group. The apoptosis and tissue necrosis were more pronounced when mice were treated with both UMI-77 and Carfilzomib (Figures 6 and 7).
Note: (A) Effect of UMI-77 on the viability of HEC-1-A and Ishikawa cells (n=3) compared with the control group. (B) Inhibitory rate (%) and CI of graded concentrations of CFZ (nM)/UMI-77 (μM) on HEC-1-A. (C) Inhibitory rate (%) and CI of graded concentrations of CFZ (nM)/UMI-77 (μM) on Ishikawa. (D) HE staining results of different groups of tumor tissues under optical microscope. (E) Inhibitory effect of CFZ/UMI-77 on endometrial cancer nude mouse models (n = 3)* P<0.05, **P<0.01.
Discussion
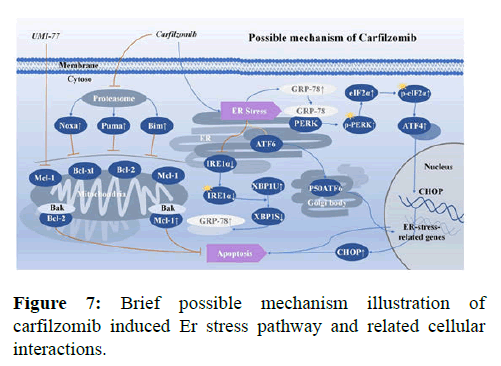
Carfilzomib induces cell apoptosis and ER stress signaling by differentially regulating the signaling pathways underlying different malignant tumors: Sustained and severe ER stress can cause cells to undergo apoptosis via at least three signaling pathways. In the PERK/ ATF4 pathway, the dissociation of PERK and GRP78 after dimerization is followed by elevated levels of PERK phosphorylation and p-PERK specific phosphorylation of eIF2α. Consequently, a large amount of p-eIF2α enables the expression of ATF4, which enters the nucleus to activate the CHOP apoptotic pathway, leading to apoptosis and expression of Mcl-1. In the ATF6 pathway, ATF6 proteins are transported to the Golgi apparatus, where they produce a 50- kDa P50ATF6-containing Hepatitis-B virus fragment, which eventually migrates to the nucleus to activate the expression of UPR target genes. Finally, XBP1 of the IFR1α-XBP1 pathway is a basic leucine zipper-type transcription factor with no transcriptional activity. IRE1α can be activated by auto-phosphorylation and possesses endonuclease activity. Activated IRE1α can cleave a 26-bp intron from the XBP1 mRNA to change its open reading frame. The alternatively spliced translational product of XBP1 can then promote the expression of UPR related genes (e.g. GRP78 and GRP94), which contain ER stress responsive elements. In this way, XBP1 attenuates ER stress signaling to restore intracellular homeostasis. In this study, markers of these signaling pathways were evaluated in response to Carfilzomib treatment, with the results suggesting that ER stress signaling in UCEC cells triggered by Carfilzomib is different from multiple myeloma and adrenocortical carcinoma. Regarding the PERK/ATF4 pathway, Carfilzomib enhanced expression of p-PERK and p-eIF2α and their downstream product ATF4. Meanwhile, this change also led to the enhancement of Mcl-1 expression. However, Carfilzomib had little effect on the transcription of ATF6, suggesting that it does not significantly affect the ATF6 pathway. Finally, in the IFR1α-XBP1 pathway, Tunicamycin caused the alternative splicing of XBP1 mRNA and produced XBP1S by endonucleases, whereas Carfilzomib did not. Carfilzomib only enhanced the levels of the unsliced precursor XBP1U. Thus, Carfilzomib can potentially inhibit the activation of the IRE1α-XBP1 pathway. These changes differ from those previously found in multiple myeloma and adrenocortical carcinoma. Thus, the origination of cells potentially an influence the part ER stresses is playing in apoptosis and response proportion of three classic pathways. Regarding apoptotic proteins, Carfilzomib brought about an accumulation of Mcl-1 and other BH3-only members in the Bcl-2 pro-apoptotic family (e.g. Bim, Puma, and Noxa) and the suppression of Bcl-2 anti-apoptotic protein, which demonstrated an underlying connection between classic ER stress and mitochondrial related apoptosis pathways. The ER stress triggered by Carfilzomib in UCEC cells was briefly illustrated (Figure 7). However, questions and limitations exist in the current research. First, present studies demonstrated that the accumulation of p-eIF2α/eIF2α/ATF4 is enhanced by the severity of ER stress. Excluding human factors, the western blotting result of p-eIF2α/eIF2α showed a decrease in 24 h group in both cell lines, and future studies should investigate the possible reason. More in vivo experiments need to be performed to confirm the changes in the three pathways discovered in vitro. The specific mechanisms underlying the differences in pathways influenced by PIs in different tumor cells require further studies, and the significance of activating these divergent ER stressing pathways remains to be illustrated. Moreover, rescue experiments, different UCEC cell lines and multisystem cancer cell lines, and animal experiments on a large scale are important focus areas for future studies.
Carfilzomib and UMI-77 induced apoptosis indicated potential chemotherapeutic management of UCEC: Sustained administration of Carfilzomib caused severe ER stress signaling in UCEC cells, which caused endogenous mitochondrial pathway-related cell apoptosis. Studies have shown that Bcl-2 family pro-apoptotic BH3-only members can stimulate the expression of apoptotic genes, Bax and Bak, to inhibit the activity of anti-apoptotic proteins. The anti-apoptotic Mcl-1 protein is a key molecule in this process demonstrated that Bortezomib activates ATF4, which up regulates Mcl-1 and leads to anti-apoptotic drug resistance. Previous studies demonstrated that down-regulation of Mcl-1 can lead to apoptosis of tumor cells and increase chemotherapy sensitivity, indicating that regulation of Mcl-1 can influence the tolerance of malignant tumor cells. In our studies, to further increase the inhibitory effect on UCEC cells, Carfilzomib was combined with a selective inhibitor of Mcl-1, UMI-77, to determine its potential chemotherapeutic function. We found an obvious coordinated effect of Carfilzomib and UMI-77 in the treatment of UCEC in vitro and in vivo. With respect to our findings, Carfilzomib, as a PI, was designed to cause up regulations of pro-apoptotic factors. The up-regulation of Mcl-1, as an anti-apoptotic factor, demonstrated the self-rescue behavior of cancer cells when treated by Carfilzomib. Moreover, UMI-77 inhibited the function of Mcl-1 to enhance the efficiency of PI treatment.
Conclusion
In summary, we treated the UCEC cells with ER stressinducing drugs and illustrated the underlying mechanism. Moreover, we found that the suppression of Mcl-1 via UMI-77 supplementation could be a solution to enhance the anti-tumor function of Carfilzomib which maintains clinical significance. Furthermore, anti-tumor target specific signaling regulators, both synthetic and those derived from natural compounds with better effectiveness, will be a necessary focus for future research.
Funding
This research was supported by research funding from the Natural Science Foundation of Shaanxi Province (CN) (Grant Number: 2021JM-035) and National Natural Science Foundation of China (Grant Number: NSFC81372534) and National Natural Science Foundation of China (Grant Number: NSFC81570192).
Acknowledgement
None.
Competing Interests
The authors declare that they have no competing interests, and all authors have confirmed its accuracy.
Availability of Data and Materials
The data used or analyzed in the study are available from the corresponding author with reasonable request.
Authors’ Contributions
• Zhaozu Feng: Methodology, Laboratory experiments, Data collection, Manuscript Drafting and Revising.
• Ke Wang: Methodology, Laboratory experiments, Data collection.
• Yunfeng Ma: Methodology, Manuscript Revising.
• Wenjuan Luo: Methodology, Manuscript Revising, Funding management.
Ethics Approval and Consent to Participate
The experimental protocol was established, according to the Laboratory animal guideline for the ethical review of animal welfare and was approved and supervised by the laboratory animal ethics committee of Xi’an Jiao tong University (No. 2016-228).
Patient Consent for Publication
No human patients were included in this study.
References
- Areeb Z, Stylli SS, Ware TM, Harris NC, Shukla L, Shayan R, et al. Inhibition of glioblastoma cell proliferation, migration and invasion by the proteasome antagonist carfilzomib. Med Oncol. 2016;33:53-60. [Crossref][Googlescholar][Indexed]
- Beauchamp EM, Platanias LC. BH3 mimetics and multi-kinase inhibition in AML. Blood. 2012;119:5947-5948. [Crossref][Googlescholar][Indexed]
- Bijnsdorp IV, Giovannetti E, Peters GJ. Analysis of drug interactions. Methods Mol Biol. 2011;731: 421-434. [Crossref][Googlescholar][Indexed]
- Burki TK. New risk loci for endometrial cancer identified. Lancet Oncol. 17:221-229. [Crossref][Googlescholar][Indexed]
- Chalmers F, Van Lith M, Sweeney B, Cain K, Bulleid NJ. Inhibition of IRE1alpha-mediated XBP1 mRNA cleavage by XBP1 reveals a novel regulatory process during the unfolded protein response. Wellcome Open Res. 2017;2:28-36. [Crossref][Googlescholar][Indexed]
- Cnop M, Toivonen S, Igoillo-Esteve M, Salpea P. Endoplasmic reticulum stress and eIF2alpha phosphorylation: The Achilles heel of pancreatic beta cells. Mol Metab. 6:1024-1039. [Crossref][Googlescholar][Indexed]
- Desantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding Sauer A, et al. Breast cancer statistics, CA Cancer J Clin. 2019;69:438-451. [Crossref][Googlescholar][Indexed]
- Fricker LD. Proteasome Inhibitor Drugs. Anun Rev Pharmacol Toxicol. 2020;60:457-476. [Crossref][Googlescholar][Indexed]
- Galimberti V, Kinor Y, Shav-Tal, Biggiogera M, Bruning A. The stress-inducible transcription factor ATF4 accumulates at specific rRNA-processing nucleolar regions after proteasome inhibition, Eur J Cell Biol. 95:389-400. [Crossref][Googlescholar][Indexed]
- Gao MG, Chen H, Wang B, Xie L, Hu Y, Kong G, et al. Therapeutic potential and functional interaction of carfilzomib and vorinostat in T-cell leukemia/lymphoma. Oncotarget. 2016;7:29102-29115. [Crossref][Googlescholar][Indexed]
- Hu J, Dang E, Menu E, De Bruyne D, Xu B, Van Camp, et al. Activation of ATF4 mediates unwanted Mcl-1 accumulation by proteasome inhibition. Blood. 2012;119:826-837. [Crossref][Googlescholar][Indexed]
- Katsnelson A. Next-generation proteasome inhibitor approved in multiple myeloma. Nat Biotechnol. 2012;30:1011-1012. [Crossref][Googlescholar][Indexed]
- Killock D. Haematological cancer: ASPIRE for unprecedented benefit with carfilzomib in MM. Nat Rev Clin Oncol. 2015;12:65. [Crossref][Googlescholar][Indexed]
- Kim KB, CM Crews. From epoxomicin to carfilzomib: chemistry, biology, and medical outcomes', Nat Prod Rep. 2013;30:600-604. [Crossref][Googlescholar][Indexed]
- Kroiss M, Sbiera S, Kendl S, Kurlbaum M, Fassnacht M. Drug Synergism of Proteasome Inhibitors and Mitotane by Complementary Activation of ER Stress in Adrenocortical Carcinoma Cells. Horm Cancer. 2016;7:345-355. [Crossref][Googlescholar][Indexed]
- Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci USA. 2003;100:9946-9951. [Crossref][Googlescholar][Indexed]
- Liu Y, Jiang YL, Zhou HH, Qiu G, Wang Y, Luo JB, et al. beta-elemene regulates endoplasmic reticulum stress to induce the apoptosis of NSCLC cells through PERK/IRE1alpha/ATF6 pathway, Biomed Pharmacother. 2017;93:490-497. [Crossref][Googlescholar][Indexed]
- Morice P, Leary A, Creutzberg C, Abu-Rustum N, Darai E.Endometrial cancer, Lancet. 2016; 387:1094-1108. [Crossref][Googlescholar][Indexed]
- Rozpedek W, Pytel D, Mucha B, Leszczynska H, Diehl JA, Majsterek I. The Role of the PERK/eIF2alpha/ATF4/CHOP Signaling Pathway in Tumor Progression During Endoplasmic Reticulum Stress. Curr Mol Med. 2016;16:533-544. [Crossref][Googlescholar][Indexed]
- Yang CS, Maliakal P, Meng X. Inhibition of carcinogenesis by tea. Annu Rev Pharmacol Toxicol. 2002;42:25-54. [Crossref][Googlescholar][Indexed]
- Zang Y, Thomas SM, Chan ET, Kirk CJ, Freilino ML, Delancey HM, et al. The next generation proteasome inhibitors carfilzomib and oprozomib activate prosurvival autophagy via induction of the unfolded protein response and ATF4. Autophagy. 2012;8:1873-1874. [Crossref][Googlescholar][Indexed]
- Zhou Y, Wang K, Zhen S, Wang R, Luo W. Carfilzomib induces G2/M cell cycle arrest in human endometrial cancer cells via upregulation of p21(Waf1/Cip1) and p27(Kip1). Taiwan J Obstet Gynecol, 2016;55(6):847-851. [Crossref][Googlescholar][Indexed]
- Ziogas DC, Terpos E, Kastritis E, Dimopoulos MA. An overview of the role of carfilzomib in the treatment of multiple myeloma. Expert Opin Pharmacother. 2017;18:1883-1897. [Crossref][Googlescholar][Indexed]




 The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.
The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.