Characterization of Escherichia coli Phylogenetic Groups Associated with Extraintestinal Infections in South Indian Population
- *Corresponding Author:
- Dr. K. Vishwas Saralaya
Department of Microbiology, Kasturba Medical College, Manipal University, Mangalore - 575 001, Karnataka, India.
E-mail: vishwassaralaya@yahoo.com
Citation Chakraborty A, Saralaya V, Adhikari P, Shenoy S, Baliga S, Hegde A. Characterization of Escherichia coli phylogenetic groups associated with extraintestinal infections in South Indian population. Ann Med Health Sci Res 2015;5:241-6.
Abstract
Background: Escherichia coli strains mainly fall into four phylogenetic groups (A, B1, B2, and D) and that virulent extra‑intestinal strains mainly belong to groups B2 and D. Aim: The aim was to determine the association between phylogenetic groups of E. coli causing extraintestinal infections (ExPEC) regarding the site of infection, expression of virulence factors, antimicrobial resistance patterns, and clinical outcome. This descriptive study was carried out in a multi‑specialty Tertiary Care Hospital. Materials and Methods: A total of 300 E. coli causing ExPEC were studied. Triplex polymerase chain reaction was used to classify the phylogenetic groups; hemolysin production was assessed on sheep blood agar and biofilm production in a microtiter plate assay. Production of extended spectrum of beta‑lactamase (ESBLs) was detected by combination disk method; AmpC was detected by AmpC disk test, Carbapenemase production was detected by modified Hodge test and metallo‑β‑lactamase by metallo‑beta‑lactamases (MBL) E‑test. Results: Of 300 isolates, 61/300 (20 %) belonged to phylogroup A, 27/300 (9%) to phylogroup B1, 104/300 (35%) were B2 and 108/300 (36%) belonged to group D, respectively. Phylogroups B2 and D were the most predominant groups in urinary tract infection and sepsis. Prognoses were better in infections with group A and B1 isolates, and relapses and death were common in infections with B2 and D. Expression of biofilm was greatest in B1 and hemolysin in group B2. Group A and B1 showed higher resistance to ciprofloxacin and were most frequent β‑lactamase (ESBL, AmpC, Carbapenemase and MBL) producers. Conclusions: Phylogenetic group B2 and D were predominant in ExPEC and exhibited least antimicrobial resistance among the groups. Resistance to multiple antibiotics was most prevalent in group A and B1. Regular monitoring of antimicrobial susceptibility in commensal strains is essential as they might transfer the property of antimicrobial resistance to pathogenic strains.
Keywords
Drug resistance, Escherichia coli, Extraintestinal infections, Polymerase chain reaction, Phylogenetic group, Virulence
Introduction
Escherichia coli strains are implicated in a large number of extraintestinal infections (ExPEC) in humans such as urinary tract infection (UTI), bacteremia, pneumonia, soft-tissue infection, and neonatal meningitis.[1] Phylogenetic analysis has shown that E. coli strains fall into four main groups (A, B1, B2, and D). It has been found that pathogenic E. coli strains causing extraintestinal infections mainly belong to group B2 and a lesser extent to group D whereas commensal strains belong to group A and B1.[2] Results of various studies have also indicated that the phylogroups A and B1 (commensal strains) exhibit an increased drug resistance pattern but possess few virulence genes whereas the phylogroups B2 and D (pathogenic strains) possess several pathogenicity associated islands and express multiple virulence factors such as adherence factors including biofilm production and high surface hydrophobicity, toxin (hemolysin and CNF) and siderophore production but are more susceptible to antibiotics.[3]
In recent years, much attention has been given to the analysis of the phylogenetic affiliation of pathogenic E. coli strains, so as to understand the sources of such ExPEC and also to limit the spread of multidrug resistance among such strains.[4] Although several studies have been reported from outside India, not much information is available regarding the phylogenetic lineage of ExPEC causing extraintestinal infections from within our country with only very few studies being reported regarding phylogenetic grouping. The aim of the present study was to assess the association between phylogenetic groups of E. coli and site of infection, phenotypic characters, antibiotic resistance, and clinical outcome, respectively.
Materials and Methods
Participants and clinical isolates
The study was conducted during the period from August 2010 to January 2012, from hospitalized patients of two Tertiary Care Hospitals, after obtaining permission by way of informed consent from patients and ethical clearance from the Kasturba Medical College, Mangalore institutional ethical committee. Sample size was determined with 55% confidence and 90% power based on previous studies. Three hundred non-repeat strains of E. coli were isolated from specimen such as urine, blood, wound swab, pus, cerebrospinal fluid, ascitic fluid, and intravascular devices, collected using standard sterile procedures from the study population. The study population included hospitalized patients of all age groups whose extraintestinal clinical samples grew E. coli and excluded those subjects who had received antimicrobial drugs during the past 1 month, who had asymptomatic UTI, polymicrobial infections and those who were discharged without treatment with antimicrobial drugs. Clinical data from the patient’s records were collected in a proforma. All patients were followed up for the period of 1 year to monitor clinical outcome. The samples were processed immediately using standard procedures. The isolates were identified based on colony morphology on Blood agar, MacConkey’s agar, Gram staining and by standard biochemical tests.[5] ExPEC isolates from blood were identified using automated biochemical system Vitek 2 (BioMerieux, France).
Biofilm production
The capacity to form biofilms was assayed in microtiter plates with minor modification as described by O’Toole and Kolter.[6] Briefly, cells were initially grown for 18 h in Trypticase soy broth (TSB) at 37°C, subsequently cultures were diluted 1:100 with fresh TSB and 200 μl were inoculated into 96 well polystyrene microtiter plates and incubated for 24 h at 37°C. After incubation, content of each well was gently removed by tapping the plates. The wells were washed four times with 200 μl of phosphate buffer saline (PBS, pH 7.2) to remove free-floating planktonic bacteria. Biofilms formed by adherent organisms in plate were fixed with Bovin’s fixative and stained with crystal violet (0.1% w/v). Excess stain was rinsed off by thorough washing with deionized water and plates were kept for drying. Optical densities (OD) of stained adherent bacteria were determined with an ELISA reader at wavelength of 630 nm. These OD values were considered as an index of bacteria adhering to surface and forming biofilms; strains with mean OD values of < 0.120 were considered as non-biofilm producers and those with mean OD values >0.120 were considered to be biofilm producers. Pseudomonas aeruginosa ATCC 27853 was used as the control organism.
Hemolysin production
Production of α-hemolysin was tested on 5% sheep blood agar. E. coli strains were inoculated onto blood agar plates, incubated overnight at 37°C and hemolysis was detected by the presence of a zone of complete lysis of the erythrocytes around the colony.[7]
Antimicrobial susceptibility testing
Antibiotic susceptibility testing was done by the modified Kirby-Bauer disk diffusion method in accordance with CLSI guidelines.[8] The antibiotic disks (HiMedia, Mumbai) used were ampicillin (10 μg), piperacillin (10 μg), piperacillin + tazobactam (100/10 μg), ceftriaxone (30 μg), cefotaxime (30 μg), ciprofloxacin (5 μg), norfloxacin (10 μg), amikacin (30 μg), gentamicin (10 μg), co-trimoxazole (1.25/23.75 μg), cefoperazone + sulbactam (75/30 μg), imipenem (IPM; 10 μg), meropenem (MRP; 10 μg), and ertapenem (ETP; 10 μg).
Screening for extended spectrum of beta-lactamase production
Isolates which were resistant to one or more third-generation cephalosporins were tested for Extended spectrum of beta-lactamase (ESBL) production by the combination disk method using, ceftazidime (30 μg) and ceftazidime/clavulanic acid (10 μg). A ≥5 mm increase in diameter of the inhibition zone of the cephalosporin-plus-clavulanate disc, when compared to the cephalosporin disc alone was interpreted as phenotypic evidence of ESBL production.[8]
Detection of AmpC production
Isolates were tested for AmpC enzyme production by AmpC disk test.[9] Briefly, a suspension of ATCC E. coli 25922 standardized to 0.5 McFarland was inoculated on the surface of Mueller–Hinton Agar (MHA) plate. A 30 μg cefoxitin disk was placed on the inoculated surface of the agar. A sterile plain disc containing Tris-ethylenediaminetetraacetic acid (EDTA) inoculated with several colonies of the test isolate was placed beside the cefoxitin disc almost touching it, with the inoculate in contact with the agar surface. The plates were incubated overnight at 35°C, aerobically. A positive test was indicated as flattening or indentation of cefoxitin inhibitory zone in the vicinity of the test disc. A negative test had an undistorted zone.
Detection of carbapenemase production
Plates of MHA were inoculated with suspensions of test strains and adjusted to a turbidity equivalent to 0.5 McFarland standard. A set of discs (HiMedia) of IPM, MRP, and ETP (10 μg each) were applied to the surface of the agar, plates were incubated overnight at 35°C aerobically, and diameters of zone of inhibition (≥23 mm indicating sensitivity, 20–22 mm indicating intermediate resistance and ≤19 mm indicating resistance) were recorded. Carbapenemase production was further confirmed by modified Hodge test.[8]
Detection of metallo-β-lactamase producers
Identification of metallo-beta-lactamases (MBL) activity was performed by two methods: A carbapenem–EDTA combined disk method and MBL E-test (HiMedia).[10] A known MBL producing isolate (PCR positive with MBL genes) was used as a positive control for all tests.
Determination of Escherichia coli phylogenetic groups
Isolates were assigned to one of the four main phylogenetic groups of E. coli (A, B1, B2, and D) by using the triplex PCR as described by Clermont et al.[11] Briefly, template DNA was obtained by boiling-lysis method. The genes chuA, yjaA and TSPE4.C2 were amplified using appropriate oligonucleotide primers.[11] The PCR was performed in a final reaction volume of 50 μl containing 750 mM Tris-HCl, 200 mM (NH4)2SO4, 2.5 mM MgCl2, 0.2 mM each of dNTP’s, 0.4 μM of each primers, 1 U of Taq DNA polymerase and 5 μl template DNA. An Eppendorf thermocycler was used for amplification. The program for amplification included a step of initial denaturation at 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 30 s and a final extension step at 72°C for 7 min. The PCR products were loaded in 2% w/v agarose gel prepared in Tris-borate–EDTA buffer at 120V for 1 h and detected by ethidium bromide staining after electrophoresis.
Statistical analysis
Chi-square test was used to find an association between the phylogroups, virulence factor genes and patient’s clinical outcome. Analysis was performed using statistical package SPSS version 17.0 (USA).
Results
A total of 300 E. coli isolates from extraintestinal sources were collected. One hundred forty-three isolates were from the medical unit, 44 from surgical, 43 from urology, 20 from oncology, 20 from gastroenterology, 13 from obstetrics and gynecology, 12 from orthopedics and 5 from pediatrics units. The primary site of infection included 159/300 (53 %) cases of UTI, 77/300 (25.6%) with bacteremia, 40/300 (13.3%) with wound infection, 19/300 (6.3%) with pneumonia, 3/300 (1%) intravascular device infection, and 2/300 (0.6%) with meningitis.
Phylogenetic analysis of isolates carried out by triplex PCR [Figure 1] indicated that 61/300 (20%) isolates belonged to Group A, 27/300 (9%) were group B1, 104/300 (35%) were group B2, and 108/300 (36%) belonged to group D. The distribution for each phylogenetic group according to gender, age group, site of infection, and outcome is summarized in Table 1. The phylogroups B2 and D occurred with greater frequency (statistically significant, P < 0.05) than A and B1 among all isolates of ExPEC without any sex disparity. Furthermore, phylogroups B2 and D were seen to occur at a far greater frequency in all age groups, when compared to A and B1. The largest number of ExPEC infections were observed in the > 60 years age group (130/300 strains; 43.3% of all infections, statistically significant, P = 0.04) indicating that this group was more susceptible than others to infection with ExPEC. UTI was the clinical entity wherein highest number of ExPEC were isolated (159/300 strains; 53% statistically significant, P = 0.03), again with phylogroups B2 and D being isolated most frequently (38.4 and 33.9% respectively, P < 0.01).
| Feature | Phylogenetic groups (n=300) | Total (%) | |||
|---|---|---|---|---|---|
| A | B1 | B2 | D | ||
| Gender | |||||
| Male | 35 | 14 | 56 | 58 | 163 (54.3) |
| Female | 26 | 13 | 48 | 50 | 137 (45.6) |
| Age | |||||
| >1 | - | 1 | 2 | 1 | 4 (1.3) |
| 1-18 | - | - | 5 | 3 | 8 (2.6) |
| 19-44 | 15 | 9 | 20 | 27 | 71 (23.6) |
| 45-59 | 18 | 10 | 32 | 27 | 87 (29) |
| >60 | 28 | 7 | 45 | 50 | 130 (43.3)# |
| Infection | |||||
| UTI | 27 | 17 | 61 | 54 | 159 (53)¥ |
| Sepsis | 16 | 7 | 27 | 27 | 77 (25.6) |
| Wound | 8 | 2 | 13 | 17 | 40 (13.3) |
| Pneumonia | 9 | - | 3 | 7 | 19 (6.3) |
| Meningitis | - | 1 | - | 1 | 2 (0.6) |
| Outcome | |||||
| Improved | 48 | 17 | 62 | 75 | 202 (67.3) |
| Re-infection | 4 | 6 | 28 | 17 | 55 (18.3) |
| Expired | 8 | 2 | 12 | 11 | 33 (11) |
| Lost to follow-up | 1 | 2 | 2 | 5 | 10 (3.3) |
| Total* n (%) | 61 (20.3) | 27 (9) | 104 (34.6)@ | 108 (36)@ | 300 (100) |
Table 1: Comparison of different phylogenetic groups of ExPEC in relation to demographical features and clinical outcome
Regarding clinical outcome, prognosis of infections with strains belonging to A and B1 phylogroups was far better than with infections due to strains from B2 and D. Seventy-eight percent of cases due to ExPEC strains belonging to A and 62.9% of cases due to infection with strains belonging to B1 were successfully treated and such ExPEC were eradicated completely with appropriate antibiotic therapy. On the other hand, higher morbidity (relapses and recurrent infections) and mortality was observed in infections due to ExPEC strains belonging to the virulent phylogroups B2 and D (37% and 26%, respectively; Table 1).
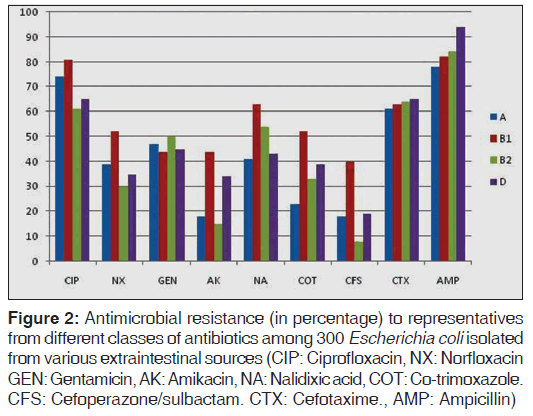
The study of phenotypic characteristics such as expression of virulence factors and antimicrobial resistance of the isolates belonging to each phylogenetic group showed that expression of biofilm production was greatest with groups B1 and B2; group B2 isolates were observed to be most frequently associated with hemolysin production (37.5%, statistically significant, P < 0.05) and 17.3% were non-lactose fermenters [Table 2]. The expression of various enzymes associated with antimicrobial resistance and resistance patterns exhibited by isolates of the four phylogenetic groups against representative antimicrobials from different classes are shown in Table 2 and Figure 2. Results of our study showed that group B2 isolates produced the least amount of beta lactamases when compared with group A, B1, and D. Resistance to ciprofloxacin, norfloxacin, co-trimoxazole, nalidixic acid and cefoperazone/sulbactam was especially prevalent among group B1 isolates and group A isolates showed comparatively higher resistance to ciprofloxacin and norfloxacin when compared with groups B2 and D. However, in case of ampicillin and cefotaxime, resistance rates were higher with group B2 and D. The presence of multi-drug resistance (resistance to three or more antibiotic groups) strains were significantly higher in group B1 isolates (statistically significant, P = 0.04). Among the four groups, A and B1 group isolates were most frequent β-lactamase (ESBL, AmpC, Carbapenemases, MBL) producers when compared to group B2 and D [Table 2].
Figure 2: Antimicrobial resistance (in percentage) to representatives from different classes of antibiotics among 300 Escherichia coli isolated from various extraintestinal sources (CIP: Ciprofloxacin, NX: Norfloxacin GEN: Gentamicin, AK: Amikacin, NA: Nalidixic acid, COT: Co-trimoxazole. CFS: Cefoperazone/sulbactam. CTX: Cefotaxime., AMP: Ampicillin)
| Phenotypic properties | Phylogenetic group (n=300) (%) | Total* | ||||
|---|---|---|---|---|---|---|
| A (n=61) | B1 (n=27) | B2 (n=104) | D (n=108) | |||
| Biofilm production | 29 (47.5) | 14 (51.8) | 51 | (49.1) | 35 (32.4) | 129 (43) |
| Hemolysis on sheep blood agar | 12 (19.6) | 4 (14.8) | 39 | (37.5) | 21 (19.4) | 76 (25.3) |
| Lactose fermentation | 58 (95.1) | 26 (96.3) | 86 | (82.7) | 91 (84.3) | 261 (87) |
| ESBL production | 42 (68.8) | 20 (74.1) | 72 | (69.3) | 78 (72.2) | 212(70.6) |
| AmpC producers | 20 (32.7) | 12 (44.4) | 23 | (22.1) | 40 (37.1) | 95 (31.6) |
| Carbapenemase producers | 5 (8.2) | 10 (37.1) | 4 | (3.8) | 10 (9.3) | 29(9.6) |
| MBL producers | 3 (4.9) | 7 (25.9) | 3 | (2.8) | 4 (3.70) | 17(5.6) |
Table 2: Expression of phenotypic properties associated with virulence and antimicrobial resistance among ExPEC strains
Discussion
Escherichia coli is emerging as an important cause of extraintestinal infections in our hospitals. These strains are seen to exhibit several virulence properties as well as a high rate of antibiotic resistance which is of major concern for management of cases. Extraintestinal pathogenic E. coli which routinely cause infections have been shown to belong to phylogroups B2 and D.[2,3] Results of our study indicated that approximately 70.6% of the E. coli isolates from our patients also belonged to phylogenetic group B2 and D which is in agreement with previous findings.[3,4] The least frequently isolated phylogenetic group in our study was group B1, which is also in accordance with similar studies done elsewhere.[4]
Our study on the prevalence of different phylogroups in various clinical entities yielded the following facts: In cases of sepsis among our study population, we found that all four phylogroups occurred with approximately equal frequency whereas in other studies, groups B2 and D have been reported to be more common.[4,12] However, in cases of UTI, B2,and D phylogroups were more common. The most common phylogroup which was responsible for relapses and/or deaths was group B2, which probably indicates the capability of strains of this group to cause persistent and severe infections.
Previous studies have indicated that phylogroups B2 and D possess more virulence properties such as biofilm formation and hemolysin secretion when compared with phylogroups A and B1 isolates.[13,14] In our study, we found group B2 consisted of a greater number of non-lactose fermenting isolates, and also more strains produced hemolysin which is considered to be a potent virulence factor essential for survival in extraintestinal sites.[3,13] Interestingly, it was group B1 isolates which expressed a higher capacity to form biofilms, a virulence factor which has been shown to be responsible for the initial step of adherence to various cell/tissue surfaces, during the process of infection.[14]
Regarding antimicrobial resistance exhibited by isolates from different phylogroups, our study results indicated that phylogroup B2 isolates were the most susceptible (statistically significant) to antibiotics than isolates belonging to the other three groups. Similar results have been obtained by other investigators, indicating that although being more virulent, the isolates of phylogroup B2 were more susceptible to antibiotics.[12] However, in our study, we found gentamicin resistance was more common in group B2 isolates. Several studies have reported a significant association of ciprofloxacin–resistant extraintestinal E. coli with phylogeny group A and ciprofloxacin susceptibility with group B2, which is similar to our study findings.[15,16]
Our study of the ability of isolates to produce antimicrobial degrading enzymes such as ESBLs, AmpC, carbapenemase, and MBLs indicated that beta lactamase producing isolates occurred with greater frequency in group A and B1 isolates when compared with B2 and D, a finding which is in accordance with other studies.[15,17] This finding indicates that strains belonging to phylogroups A and B1 were carrying more resistance properties than the strains belonging to phylogroups B2 and D. By this finding, it may be assumed that in our population a large number of people act as a reservoir for such resistant strains. This may be due to inappropriate antibiotic use in both animals and humans.
Conclusion
Results of our study indicate that pathogenic strains belonging to phylogenetic groups B2 and D express more virulent properties but were seen to be more susceptible to antibiotics than phylogroups A and B1 isolates. This finding indicates a need for regular monitoring of antimicrobial susceptibility in commensal strains as these strains might transfer the property of antimicrobial resistance to pathogenic strains. There is also an urgent need to undertake measures which would prevent the development of multidrug resistance among commensal strains such as reducing inappropriate antibiotic treatment, the overuse of antimicrobials in the agricultural industry which might contribute to increased numbers of multidrug resistant strains. Clearly, antimicrobial resistance may lead to decreased therapeutic outcomes in the near future.
Acknowledgements
We would like to thank API Karnataka for their financial support and Manipal University for permitting us to conduct the study.
References
- Russo TA, Johnson JR. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J Infect Dis 2000;181:1753-4.
- Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, et al. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun 1999;67:546-53.
- Smith JL, Fratamico PM, Gunther NW. Extraintestinal pathogenic Escherichia coli. Foodborne Pathog Dis 2007;4:134-63.
- Bukh AS, Schønheyder HC, Emmersen JM, Søgaard M, Bastholm S, Roslev P. Escherichia coli phylogenetic groups are associated with site of infection and level of antibiotic resistance in community-acquired bacteraemia: A 10 year population-based study in Denmark. J Antimicrob Chemother 2009;64:163-8.
- Crichton PB. Enterobacteriaceae: Escherichia, Klebsiella, Proteus and other genera. In: Collee JG, Fraser AG, Marmion BP, Siminons A, editors. Mackie and McCartney Practical Medical Microbiology. 14th ed. New York: Churchill Livingston; 1996. p. 361-4.
- O’Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple,convergent signalling pathways: A genetic analysis. Mol Microbiol 1998;28:449-61.
- Rijavec M, Müller-Premru M, Zakotnik B, Zgur-Bertok D.Virulence factors and biofilm production among Escherichia coli strains causing bacteraemia of urinary tract origin. J Med Microbiol 2008;57:1329-34.
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. CLSI M100-S20U. Update June, 2010. Wayne, PA: Clinical and Laboratory Standards Institute; 2010.
- Black JA, Moland ES, Thomson KS. AmpC disk test for detection of plasmid-mediated AmpC beta-lactamases in E n t e r o b a c t e r i a c e a e lacking chromosomal AmpC beta-lactamases. J Clin Microbiol 2005;43:3110-3.
- Yan JJ, Wu JJ, Tsai SH, Chuang CL. Comparison of the double-disk, combined disk, and Etest methods for detecting metallo-beta-lactamases in gram-negative bacilli. Diagn Microbiol Infect Dis 2004;49:5-11.
- Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 2000;66:4555-8.
- Jaure×guy F, Carbonnelle E, Bonacorsi S, Clec’h C, Casassus P, Bingen E, et al. Host and bacterial determinants of initial severity and outcome of Escherichia coli sepsis. Clin Microbiol Infect 2007;13:854-62.
- Jakobsen L, Spangholm DJ, Pedersen K, Jensen LB, Emborg HD, Agersø Y, et al. Broiler chickens, broiler chicken meat, pigs and pork as sources of ExPEC related virulence genes and resistance in Escherichia coli isolates from community-dwelling humans and UTI patients. Int J Food Microbiol 2010;142:264-72.
- Soto SM, Smithson A, Martinez JA, Horcajada JP, Mensa J, Vila J. Biofilm formation in uropathogenic E. coli strains: Relationship with prostatitis, urovirulence factors and antimicrobial resistance. J Urol 2007;177:365-8.
- Johnson JR, Kuskowski MA, Owens K, Gajewski A, Winokur PL. Phylogenetic origin and virulence genotype in relation to resistance to fluoroquinolones and/or extended-spectrum cephalosporins and cephamycins among Escherichia coli isolates from animals and humans. J Infect Dis2003;188:759-68.
- JohnsonJR, KuskowskiMA, GajewskiA, SahmDF, KarlowskyJA. Virulence characteristics and phylogenetic background of multidrug-resistant and antimicrobial-susceptible clinical isolates of Escherichia coli from across the United States, 2000-2001. J Infect Dis 2004;190:1739-44.
- Corvec S, Prodhomme A, Giraudeau C, Dauvergne S, Reynaud A, Caroff N. Most Escherichia coli strains overproducing chromosomal AmpC beta-lactamase belong to phylogenetic group A. J Antimicrob Chemother 2007;60:872-6.




 The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.
The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.