Chloroquine Sensitivity and Prevalence of Chloroquine-resistant Genes pfcrt and pfmdr-1 in Western Kenya after Two Decades of Chloroquine Withdrawal
2 Department of Biochemistry and Biotechnology, Kenyatta University, P.O. Box 43844-00100, Nairobi, Kenya
3 Research Officer and Head of Malaria Section, Centre for Research Biotechnology and Development, Kenya Medical Research Institute, P.O. Box Nairobi, Kenya
Citation: Kishoyian G, et al. Chloroquine Sensitivity and Prevalence of Chloroquine-resistant Genes pfcrt and pfmdr-1 in Western Kenya after Two Decades of Chloroquine Withdrawal. Ann Med Health Sci Res. 2018;8:331-335
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Background: The widespread of chloroquine-resistant Plasmodium falciparum malaria in Kenya lead to the official withdrawal of chloroquine (CQ) drug by the Government as a first-line treatment of uncomplicated P. falciparum malaria in 1999. This study aimed to investigate CQ resistant in Kenya after its withdrawal. Methods: Blood samples were collected on 3M Whatman from patients infected with P. falciparum from Chulaimbo Sub-District Hospital. DNA was extracted using Chelex method for Polymerase chain reaction (PCR) amplification of pfcrt and pfmdr-1 genes. PCR--restrictive fragment length polymorphism was performed using enzymes Apol1 and Afl3 targeting point mutation of K76T and N86Y in pfcrt and pfmdr-1 respectively. The PCR products were sequenced, genotyped and further analyzed for amino acid changes in these codons. Chi square tests were used to test the significance of the findings against previous prevalence results. Results: The frequency of pfcrt wild-type codon K76 (chloroquine sensitive) stood at 76.3% while the mutant type codon 76T (chloroquine resistant) at 23.7%. Likewise, the frequency of the pfmdr-1 wild-type codon N86 was 82.9% (chloroquine sensitive) and mutant codon 86Y (chloroquine resistant) was 17.1%. Conclusion: This study observed that there is a significant (χ2 = 170.1, df=2, p<0.05) increase in CQ sensitivity in 2015 compared to 1999 when it was withdrawn indicating that the prevalence and patterns of point mutations of pfcrt (K76T) and pfmdr-1 (N86Y) is decreasing. This further suggests that CQ-sensitive P. falciparum strains are gradually returning after the withdrawal of CQ from the Kenyan population.
Keywords
Plasmodium falciparum; Mutation; Chloroquine resistant; pfcrt; pfmdr-1
Introduction
Malaria is a leading infectious parasitic disease globally with approximately a million deaths mostly in children under five years annually. [1,2] The most affected continent in the world is Africa where approximately 90% of deaths occur every year Kiarie. [3] In Kenya, it is the major cause of morbidity and mortality with 70% population at the risk of the disease as explained by the Ministry of Health (MoH). [4] However, according to the Kenya Malaria Indicator Survey (KMIS), [5] the malaria burden in Kenya is not evenly distributed with lake region and coast being endemic areas.
The rapid spread of malaria and development of resistant to chloroquine (CQ) against uncomplicated cases of malaria is a major concern in the treatment, control, and eradication of the disease worldwide. [6-8] In Africa, [9] Malawi was the first country to replace CQ with sulfadoxinepyrimethamine (SP) as the first line in the treatment for uncomplicated malaria followed by Uganda and Tanzania in 1993. [10,11] In Kenya, the first case of CQ resistant was reported in 1977. [12] In 1993, the resistance levels had reached an alarming rate of 70% [3]. In 1998, the Kenyan Ministry of Health replaces CQ with SP. [13] In 2004, SP was replaced with AL as the first line drug in the treatment of uncomplicated malaria in Kenya. [9,14]
Though microscopy is the standard tool, molecular markers have been found as a useful tool in monitoring drug-resistant. [15-17] Additionally, previous studies [2,7,9] have implicated molecular markers Plasmodium falciparum chloroquine-resistant transporter gene (pfcrt) and Plasmodium falciparum multidrug-resistant (pfmdr-1) genes as CQ resistant contributors. [8,18] Other studies have also linked high frequency of pfcrt (K76T) and pfmdr-1 (N86Y) to CQ resistant and concluded that Single Nucleotide Polymorphism (SNPs) can be used in predicting emerging resistance among commonly used CQ. [19,20] However, since point mutations from the genes result in the substitution of various amino acids changes at different positions of the codons associated with CQ resistance, monitoring point mutations is of paramount importance in the control and management of ant-malarial drugs. The purpose of this study was to find out the current frequency of pfcrt and pfmdr-1 gene mutation in the study population.
Materials and Methods
Study site
This study was carried out in Chulaimbo Sub-District Hospital in Kisumu County in western Kenya.
Study population
This included children between the age of 6 and 60 months visiting the outpatient clinic at the Hospital.
Sample collection
Sample collection was done in the month of May to November 2015. Prior to treatment, blood from a finger prick was collected and placed on filter paper for DNA extraction. The paper was air dried, labeled and transported to Kenya Medical Research Institute Nairobi, genotype analysis.
Sampling design
This was a cohort study.
Sample size
The equation used was by Lwanga and Lemeshow. [21]
n =Z2 p (1-P)/ e2
Where;
Z = Standard normal deviation of the required confidence.
n = The desired sample size
p = Proportion in the target population estimated to have characteristic being measured. According to KIAS (2010), The prevalence of malaria stands at 38% or 0.38.
e = The level of statistical significance set.
Therefore, substituting Z for 1.96, 0.38 for P and 0.1 for e, then the minimum sample size was
1.962 × 0.38 (1-0.38) / 0.12 = 90. However, only 76 whose guardian agreed to complete the follow up to 28th day after treatment with Artemether-lumefantrine.
Deoxyribonucleic acid (DNA) extraction and gene amplification
Pfcrt and Pfmdr-1 gense amplification
The extraction of DNA was processed using the Chelex method as described by Warhurst et al. [22] while the Pfcrt and Pfmdr-1 genes were amplified using a gene Amp PCR system 9700 machine. For pfcrt gene, Polymerase Chain Reaction (PCR) amplification was processed as follows; in a final reaction volume of 30 microliters (μl), the outer PCR consisted of 1x PCR buffer 1.5 Mm MgCl2, 200 μl of dNTP mixture, 100 nm each of CRTA1 (5’GCGCGCGCATGGCTCACGTTTAGGTGGAG3’) and CRTB1 (5’GGGCCCGGCGGATGTTACAAAACTATAGTTACC3’) and 0.3 μl of the enzyme Taq polymerase (Bio line, 5 U/μl). The PCR conditions were: 94°C for 3 minutes, 94°C for 30 seconds, 56°C for 30s, 60°C for 60s, 40 cycles and the final extension at 60oc for 3 minutes and then held at 4°C. From the final volume of 30 μl, the nested PCR consisted of 1μl of the outer PCR products, 1x PCR product, (Roche) 1.5 Mm MgCl2, 200 μM (deoxynucleotide triphosphate) dNTP mixtures, 100 nm each of CRTD1 (5’TGTGCTCATGTTTAAACTT3’) and CRTD2 (5’CAAAACTATAGTTACCAATTTTG3’) and 0.3 μl of Taq polymerase (Bioline, 5U/μl).
The PCR amplification for Pfmdr-1 gene was processed as follows: in an approximate reaction volume of 30 μl, the outer PCR consisted of 1x PCR buffer manufactured by Roche, 1.5 Mm MgCl2, 200 μM dNTP mix, 100 nm each MDR/A1 (5’GCGCGCGTTGAACAAAAAGAGTACCGCTG3’) and MDR/ A3 (5’GGGCCCTCGTACCAATTCCTGAACTCAC3’) and 0.3 μl of Taq polymerase 5 U/μl manufactured by Bioline. The total mixtures were topped up to the approximate volume of 50 μl with nuclear-free PCR water. The conditions for the PCR were as follows; 94°C for 3 minutes, 94°C for 1 minute, 72°C for 1 minute, 40 cycles with the final extension at 72°C for 3 minutes the halted at 4°C. In approximately 30 μl final volume, the nested PCR consisted of 1 μl of outer PCR, 1x PCR buffer manufactured by Roche, 1.5 Mm MgCl2, 200 μl of dNTP each of MDR/A2 (5’TTTACCGTTTAAATGTTTACCTGC3’) and MDR/A4 (5’CCATCTTGATAAAAAACATTTCT3’) and 0.3 μl of the Taq polymerase manufactured by Bioline 5 U/μ. This was followed by topping up the mixture to a volume of approximately 50 μl with nuclease-free PCR water. After that, the conditions were set as follows; 94°C for 3 minutes, 94°C for 30 s, 45°C for 1 minute, 72°C for 1 minute, 40 cycles while final extension at 72°C for 3 minutes and finally at 4°C.
Restrictive digest of Pfcrt and Pfmdr-1 genes
The allele-specific restriction analysis for the Pfcrt gene was done using restriction endonuclease Apo1 as per the New England Bio Labs protocol. The allele for specific restriction analysis for Pfmdr-1 gene was processed using restriction endonuclease AflIII manufactured by the New England Bio Labs. In an approximate volume of 20 μl, 1.5 μl of 10x buffer 3, 0.15 μl of bovine serum antigen, 8 μl of amplified DNA, and 0.5 μl of AflIII of the restrictive endonuclease were added to 9.85 μl of nuclease-free PCR water. This was then incubated for 14 hours at 50°C without agitation while the digested samples were analyzed using 1.5% agarose gel manufactured by Bioline in TAE buffer and process as for the pfcrt gene.
Results
Demographic characteristics
Of the study population, the mean was 8.30 ± 6.919 while the mean weight was 6.55±3.390. According to gender, males were 43 (56.6%) while 33 (43.4%) were females. The geometric mean of microscopic parasite count on day 0 (before treatment) was 133,960 parasites/μl (95% CI: 2152.66-4545.65), day 1 was 1,664 parasites/μl (95% CI: 540-2788) while on day 2, there were no parasites up to day 28 among all the children
Prevalence of pfcrt and pfmdr-1 mutants
In addition to CQ resistant analysis based on the presence of K76T mutation in Pfcrt gene, CQ resistant analyzed using Restrictive Length Fragment Polymorphism where D6 and W2 clones were used as positive and negative controls respectively. The D6 control gave two fragments of 99 and 46 bp as expected while the W2 was resistant to digestion. These controls were characterized as either mutant or wildtype.
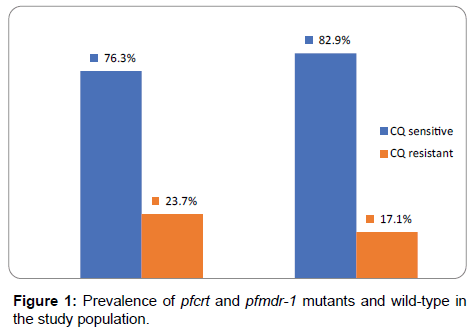
Using this characterization, out of the 76 samples digested with ApoI enzyme, 76.3% (95% CI: 0.668–0.833) were CQ sensitive, while 23.7% (95% CI: 0.167–0.332) were resistant to digestion and thus CQ resistant as shown in Figure 1. In the case of mixed samples harboring both resistant and sensitive clones, ApoI digestion was expected to yield three fragments 145, 99 and 46 bp in length. None of the samples yielded this type of banding pattern thus no mixed samples were identified.
For pfmdr-1, all the samples gave the expected 560 bp fragment. Digestion was done with Afl3 restriction enzyme using DD2 and 3D7 as positive and negative controls respectively. Digestion of DD2 clone yielded 2 fragments of 227 and 233 base pair fragments as expected, while the 3D7 clone was not sensitive to digestion. With the same criterion, field samples subjected to digestion were classified as either mutant or wild-type. Of the 76 samples digested [Figure 1] 17.1% (95% CI 0.109-0.225) were restricted (mutants) while 82.9% (95% CI 0.745-0.891) were not digested (non-mutants) thus sensitive to CQ.
Discussion
The current study was done to monitor CQ resistant based on the detection of point mutations in the pfcrt and pfmdr-1 genes. The results showed that the prevalence of pfcrt K76T and the pfmdr-1 N86Y mutation was 23.7% and 13.1% respectively. Compared to 1999 when the prevalence of the mutant pfcrt allele stood at approximately 100% indicating total resistance to CQ in Kenya, the current study recorded a significant decline in the frequency of mutant alleles of pfcrt. [3] This conflicts with a similar study in Southern India where the prevalence of K76T mutation was approximately 40%. [20] However, in the same study, the prevalence of pfmdr-1 mutant was similar with 13.33% observed in the current study.
Several studies have observed that resistant to CQ is associated with point mutation in the Pfmdr-1 gene pointing to a change of an amino acid at codon 86, 184, 1034, 1042 and 1246. [23] However, conflicting findings have been reported in several studies. For example, in Southeast Asian, CQ resistant to P. falciparum have the replacement of the amino acid at codon 86 from Asparagine to Tyrosine (N86Y) [23] while from South American isolates mutational changes occurred at codon 184, 1034, 1042 and 1246. Out t of these, codon 86 seems to be the main dominating influence since it is involved in the substrate specificity of the gene product (P- glycoprotein) and consequently may alter the conveyance movement of the protein. [24,25]
The decreasing frequency for the pfcrt K76T mutation in the current study is consistent with several previous reports from other malariaendemic regions in Africa. For example, in Ghana a decline in frequency of the pfcrt K76T mutation between 88% to 56% from 2005-2010 was observed [7] while in Malawi and Kenya, a reduction from 85% to 13% between 1993 and 2000 and from 95% to 60% between 1993-2006 was observed respectively. [7] The similar decrease in mutant pfcrt was reported in several countries such as Senegal and Mozambique. However, the prevalence of pfcrt mutant 76T varies in the various parts of Kenya. For example, in the present study, the prevalence was 23.7% while studies were done in Kilifi, Mwea, Coastal, and Western Kenya, the prevalence was 63%, 94%, 63%, and 27.6% respectively. These conflicting observations could be due in part to inadequate enforcement on the ban of CQ and that CQ is still in use after its official withdrawal from Kenya. [3]
However, as to whether the possibility of CQ-sensitive P. falciparum returning in malaria-endemic regions after the withdrawal of CQselective pressure is of great interest for malaria control. Several studies have observed that pfcrt 76T mutation has been considered as the most reliable marker of CQ-resistant. [7] In the present study, the high frequency (76.3%) of the wild-type allele of pfcrt K76 was observed in P. falciparum isolates from the study population and can be used as a marker of CQ resistant. However, a study in Chhattisgarh in Central India observed a 22% wild-type CQ sensitive strain of pfcrt gene codon K76T in the study population. [26] This finding was found to collaborate with recent reports in other parts of India which observed that after 6 years of CQ withdrawal the prevalence was between 20- 40%. [26] However, these studies contradict the current study that observed a 76.3% of wild-type P. falciparum CQ-sensitive. However, despite this contrast differences, both observations appear to agree that though the level of sensitivity is different in different part of the world, CQ-sensitive P. falciparum strains are slowly returning after CQ withdrawal.
The withdrawal of CQ from most African countries, for example, Mozambique led to the rapid recovery of CQ sensitive P. falciparum increasing the limit fitness cost that accompanied CQ resistant. [27] However, the rapid recovery of CQ sensitive strains was not observed within the same time frame in other disease-endemic countries such as Ghana and Uganda after chloroquine withdrawal. [27] For example, a study in Burkina Faso observed that the pfcrt 76T mutation which is the main determinant of CQ resistant there was a significant decrease of its prevalence as compared to previous studies in Burkina Faso with a similarly dramatic decrease was also reported in Malawi. [6] These observations agree with the current study which observed a continuous decrease of mutant 76T from 100% in 1999 to 23.7% in 2015. This “chemo-reversion” can be due to the rapid expansion of susceptible parasites strains that survive in semi-immune hosts during the period of high chloroquine drug pressure and this phenomenon may be accelerated in areas where artemether-lumefantrine in currently in use. [28]
In Nigeria, a study was done to find out the prevalence of pfcrt and pfmdr-1 observed that N86 allele was 47.9% while 86Y was 28.3%, K76 was 17.4% while the resistant allele 76T was 12.4%. [29] This improvement after CQ withdrawal agrees with the present study where the wild-types of continuing to show remarkable success and that CQ can be used to treat malaria in the near future since pfcrt K76 and pfmdr-1 N86 do not confer resistant to CQ. [29] A subsequent study conducted in Ethiopia found that a large proportion of the study population (81.4%) carried the CQ susceptible pfcrt K76 allele. [30] However, these results contradict a previous study in the same country with a report of 100% frequency of the mutant 76T. The current study where pfcrt K76 stand at 76.3% is therefore in line with 81.4% from Ethiopia. This emerging development is important since CQ is cheap, safe, well tolerated, long-lasting and easy to prepare molecule. [30]
A study done in Kenya on the trends of established genotypes associated with tolerance to clinically important antimalarial drugs for the last two decades observed that there was a significant increase in the prevalence of pfcrt K76 wild-type allele between 1995 and 2013 from 38% to 81.7%. [31] In the same study, there was also a significant increase in the prevalence of the wild-type pfmdr-1 N86 from 14.6% in 1995 to 66% in 2013 and a corresponding decline of the mutant pfmdr-1 86Y from 43.6% to 0% in 19 years. However, the present study has observed that there is an increasing in the prevalence of the pfcrt K76T wild-type allele (19.1% in 2006 to 76.3% in 2015) and a decreasing trend for the pfcrt K76T mutant. This decreasing trend for pfcrt K76T mutation is consistent with several studies observed in Tanzania and Madagascar. [7]
In addition, the decline in the prevalence of pfmdr-1 mutation (17.1%) is lower than that of pfcrt gene mutation (23.7%) among the study population. This observation is similar to other reports describing pfmdr-1 and pfcrt polymorphism in P. falciparum isolates collected in Malawi. [7] These findings suggest that pfmdr-1 mutation may be less deleterious to parasite fitness than the pfcrt mutation even though pfmdr-1 mutation can modulate the level of CQ-resistance. These results from different studies highlight the importance of continuous surveillance using molecular markers as indicators of the therapeutic efficacy of antimalarial especially during the changes of malaria treatment policy.
In the present study, the swift from CQ-resistant to CQ-sensitive strains indicate that the replacement of CQ with Artemisinin Combination Therapy (ACT) for the treatment of P. falciparum may have resulted into a significant decline of pfcrt and pfmdr-1 mutation or CQ-resistant strains in the country. The return to CQ-sensitive could also be attributed to the emergence of susceptible strains or a new sweep in Africa. [7] Whether or not the CQ-resistant resurgent is due to the expansion of surviving CQ-reservoir populations or back mutations in the CQ-resistant allele in the study population, more studies are necessary. In addition, the possibility of using CQ in future as a combination therapy with another short acting drug with different pharmacodynamics profile will be an additional antimalarial option.
Conclusion
The current study has indicated an encouraging outcome in the prevalence of CQS parasites in the study area. This could be attributed to the removal of CQ drug from the hospitals and pharmacies resulting in the reduction of drug pressure to sustain CQR P. falciparum strains. This study concludes that the tremendous results from this study suggest the P. falciparum resistance to CQ is progressively waning which is beneficial for drug policymakers in Kenya as CQ is cheaper and can have the potential of being incorporated in future with another anti-malarial drug. Finally, further research should pay attention to cheap and cost-effective methods for continues monitoring of drug sensitivity using molecular markers with methods that are accurate and precise especially in limited-resource countries.
Strength of the Study
This study has produced encouraging results on the resistant genes prevalence of pfcrt and pfmdr-1. Though they are higher than the WHO standard of 10% threshold of drug-resistant before banning the drug, the results form a baseline study for more studies to be carried out in other malaria prone-regions of Kenya.
Limitation of the Study
The findings obtained cannot be generalized because the sample size was from a small geographical area of Kenya. In addition, the study recruited a small number of respondents and only those parents consented were included in the study. However, the study was able to achieve its objectives of determining the status of chloroquine sensitivity and prevalence of CQ-resistant genes by use of molecular markers.
Authors’ Contribution
GK came up with the study design, collected samples from the study subjects, contributed to data analysis and drafted the manuscript. EN, GO, FK and GK critically reviewed and revised the manuscript for the important intellectual content of the final manuscript. All authors read and approved the final version of the manuscript.
Availability of Data and Materials
The raw data is available on request by the editor of the publishing journal.
Acknowledgment
My appreciation is for all the patients and their mothers for participating in the study, KEMRI management for allowing me to analyze the samples in the Laboratories especially malaria laboratory staffs.
Funding Statement
Funding was partly self-sponsored and partly sponsored and by National Council of Science Technology and Innovation (NACOSTI) Ref No. 5th call 2013/2014.
Ethical considerations
The study was accepted by the Scientific and Ethical Unit of the Kenya Medical Research Institute.
Consent to Participate and Publish
Consent was obtained from mothers of the participating children and also agreed that the results can be published provided the names of the children should not appear anywhere.
Conflict of Interest
The authors disclose that they have no conflicts of interest.
REFERENCES
- Omer OA, Tahir AE, Abdelwahid MO, Gasmelseed AM, Elwali EM. Co-existence of dihydrofolate reductase (dhfr 108) gene with Plasmodium falciparum chloroquine resistance transporter gene (pfcrt T76) in P. falciparum isolates from Gezira State, Central Sudan. The Open Tropical Med. J. 2010; 3:15-17
- WHO/HTM/MAL. RBM, UNICEF, World Health Organization, Geneva, Switzerland. 2005.
- Kiarie WC, Wangai L, Agola E, Kimani FT, Hungu C. Chloroquine sensitivity: Diminished prevalence of chloroquine-resistant gene marker pfcrt-76 13 years after cessation of chloroquine use in Msambweni, Kenya. Malar J. 2015;14:328.
- Ministry of Health (MOH). Kenya malaria indicator survey 2010-2014.
- Kenya National Bureau of Statistics. Kenya National Malaria Indicator Survey 2015.
- Some AF, Sorgho H, Zongo I, Bazie T, Nikiema T, Sawadogo A, et al. Polymorphisms in K13, pfcrt, pfmdr-1, pfdhfr and pfdhps in parasites isolated from symptomatic malaria patients in Burkina Faso. Parasite. 2016;23:60.
- Zhong D, Afrane Y, Githeko A, Cui L, Menge DM, Yan G. Molecular epidemiology of drug-resistant malaria in western Kenya. BMC Infectious Diseases. 2008;8:105.
- Huang B, Qi W, Changsheng D, Wang TY, Shiguang H, Yajun L, et al. Prevalence of pfcrt and pfmdr-1 mutation in Plasmodium falciparum isolate from Grande Comore island after withdrawal of chloroquine. Malar J. 2016;15:414.
- Lim P, Alker A, Khim N, Shar NK, Incardona S, Doung S, et al. Pfmdr-1 copy number variant and artemisinin derivatives combination therapy failure in falciparum malaria in Cambodia, Malar J. 2009;8:11.
- Wangai LN, Muurira G, Omar S, Magoma G, Kimani FT, Mwangi JM, et al. Molecular screening for Plasmodium falciparum resistance markers for Artemisinins in Mbita, Kenya. Afric. J. of Clin. Exper. Microbiol. 2011;12:106-110.
- Abugri J, Ansah F, Asante KP, Opoku CN, Amenga-Etego AL, Awandare GA. Prevalence of chloroquine and antifolate drug-resistant alleles in Plasmodium falciparum clinical isolates from three areas in Ghana. AAS Open Research. 2018;1:1
- Phyo A, Nkhoma S, Stepniewska K, Ashley E, Nair S, McGready R. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet. 2012;379:1960-1966.
- Wallems TE, Plowe CV. Chloroquine-resistant malaria. J. Infect Dis. 2001;184:770-776.
- Thomsen TT, Ishengoma DS, Mmbando BP, Lusingu JP, Vestergaard LS, Theander TG. Prevalence of Single Nucleotide Polymorphisms in the Plasmodium falciparum multidrug resistance gene (pfmdr-1) in Korongwe District in Tanzania before and after the introduction of artemisinin-based combination therapy. Am. J. of Trop. Med. and Hyg. 2011;85:6:979-983.
- Ministry of Health. Kenya annual malaria report. Epidemiology of malaria in Kenya. 2013;1-2.
- Isozumi R, Uemura H, Kimata I, Ichinose Y, Logedi J, Omar AH. The novel mutation in K13 propeller gene of artemisinin-resistant Plasmodium falciparum. Emerg. Infect. Dis. J. 2015;21:490-492.
- Ouattara A, Kone A, Adams M, Fofana B, Maiga AW, Hampton S. et al. Polymorphisms in the K13-propeller gene in artemisinin-susceptible Plasmodium falciparum parasite from Bouguola-Hameau and Bandiagara, Mali. Am. J. of Trop. Med. and Hyg. 2015;92:1202-1206.
- Jutta M, Thomas AS, Hastings IM, Ivo M, Albert S, Olive O, et al. Plasmodium falciparum resistance to anti-malarial drugs in Papua New Guinea: evaluation of a community-based approach for the molecular monitoring of resistance. Malar J. 2010;9:8.
- Mohon AN, Alam MS, Bayer AG, Folefoc A, Shahinas D, Haque R, et al. Mutation in Plasmodium falciparum K13 propeller gene from Bangladesh (2009-2013). Malar J. 2014;13:431
- Nkhoma S, Nair S, Mukaka M, Molyneux ME, Ward SA, Anderson, TJC. Parasites bearing a single copy of the multi-drug resistant gene (pfmdr-1) with wild-type SNPs predominate amongst Plasmodium falciparum isolates from Malawi. Acta Trop. 2009.
- Lwanga SK, Lameshow S. Sample size in health studies: a practical manual. 1991.
- Warhurst Warhurst DC, Awad El Kariem FM, Miles MA. Simplified preparation of malarial blood samples for polymerase chain reaction. Lancet. 1991;337:303-304.
- Hiasindh AA, Sindhusuta D, Subhash CP, Sanghamitra P. Sequence analysis of pfcrt and pfmdr-1 genes and its association with chloroquine resistance in Southeast Indian Plasmodium falciparum isolates. Genome data, 2016;8:85-90.
- Shrivatsava KS, Ravi KG, Jagdish M, Mohan LD. Correlation of molecular markers, Pfmdr-1-N86Yand Pfcrt-K76T with in vitro chloroquine-resistant Plasmodium falciparum, isolated in the malaria endemic states of Assam and Arunachal Pradesh, Northeast India. Plos One. 2014;9:8.
- Ibraheem ZO, Abd Majid R, Noor MN, Sedik MH, Bashir R. Role of different pfcrt and pfmdr-1 mutation in conferring resistant to antimalarial drugs in Plasmodium falciparum. Malar Res. and Treat. 2014.
- Priyanka P, Praveen KB, Devendra B, Nazia AA, Rajive KR, Pradyumna KM, et al. Prevalence of mutations linked to antimalarial resistance in Plasmodium falciparum from Chhattisgarh, Central India: A malaria elimination point of view. Sci Rep 2017;7:16690.
- Kwame KA, Boampong JN, Duah NO, Foakwah RA, Sengal R, Quashie NB. Synergism between Pfcrt and Pfmdr-1 genes could account for the slow recovery of chloroquine- sensitive Plasmodium falciparum strains in Ghana after chloroquine withdrawal. Journal of Infection and Public Health. 2017;10:110-119.
- Thomsen TT, Madsen LB, Hansson HH, Tomas EV, Charlwood D, Bygbjerg IC, et al. Rapid selection of P. falciparum chloroquine-resistant transporter gene and multidrug-resistant gene-1 haplotypes associated with past chloroquine and present artemether-lumefantrine use in Inhambane District, South Mozambique. Am J. of Trop Med and Hgyn. 2013;88:536-541.
- Muhammad RH, Nock HI, Ndams I, George JB, Yusuf D. Distribution of pfmdr-1 and pfcrt chloroquine drug resistance alleles in north-western Nigeria. Malar Journal. 2017;8:15.
- Mekonnen SK, Aseffa A, Berhe N, Teklehaymanot T, Clouse RM, Gedru T, et al. Return of chloroquine-sensitive Plasmodium falciparum parasites and emergence of chloroquine-resistant Plasmodium vivax in Ethiopia. Malar Journal. 2014;13:244
- Okombo J, Kamau AW, Marsh K, Sutherland JC, Ochola-oyier LI. Temporal trends in the prevalence of Plasmodium falciparum drug resistance alleles over two decades of changing antimalarial policy in coastal Kenya. International Journal of Parasitology: Drug and Drug- Resistant. 2014;4:152-163.




 The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.
The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.