Heavy Metal Tolerance Pattern of Textile Dye Degrading Native Bacteria: A Bioremediation Viewpoint
2 Department of Pharmacy, Noakhali Science and Technology University, Noakhali-3814, Bangladesh
Citation: Tarequl Islam. Heavy Metal Tolerance Pattern of Textile Dye Degrading Native Bacteria: A Bioremediation Viewpoint. Ann Med Health Sci Res. 2017; 7: 67-73
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Background: Textile Industries are the major sector for social and economic perspective in Bangladesh that discharge huge number of dye stuff containing recalcitrant compounds, pigments and dye etc. into the water. Aim: The aim of this study is to isolate and identify the textile dye degrading bacteria from textile effluents and evaluation of their heavy metals resistant pattern. Methods and Materials: All the samples were collected from 8 distinct dye industry related places in Narayanganj District of Bangladesh. The isolated bacteria was characterized and identified by using cultural and biochemical techniques. The decolorization of dye was measured using spectrophotometer and the MIC of isolated bacteria against selected heavy metals’ salts was also determined by serial dilution method. Only 12 bacterial isolates of 6 species were isolated and identified based on Cultural and biochemical tests. One isolates of each species was selected for this study. Results: Pseudomonas spp. was the most effective decolorizers of textiles azo dyes than others bacterial species. All the isolates showed MIC against selected heavy metals ranging from 350-550 μg/ml where Bacillus spp., Micrococcus spp., and Pseudomonas spp. were found the most efficient heavy metal remover. Conclusion: These results signify that bacterial isolates that reside in industrial effluents could effectively be used in development of alternative and eco-friendly method for decolourization of textile dyes and removal of heavy metals from contaminated sites.
Keywords
Textile dye; MIC; Pseudomonas spp; Bacillus spp; Micrococcus spp
Introduction
Textile and paper industries are the head of Bangladesh economy but they are becoming problematic day by day because these industries release large amount of untreated or improperly treated discharge into the environment. During dyeing processes, more than 15% dye release in the discharge that don’t bind to the fiber. [1]. Azo dye is the main constituent of the textile and paper dyes that hold almost 70% of these dyes. [2]. These dyes are mostly used in the textile industry for their extensive variety of color shades and brilliant colors. Textile dyes pollute the environment heavily and contribute to different diseases. [3-5]. Azo dyes are recalcitrant xenobiotics. [6,7]. Most of these compounds are carcinogenic or contain mutagenic properties. 90% of textile dyes could remain unaffected by conventional treatment methods and discharged into the environment, [3] so to be environmentally friendly it is very important to treat these textile dyes before discharge. Bacteria can be an alternative to remove the azo dyes from the environment as many dye degrading bacteria have been identified and characterized. But their degradation capability depends on some abiotic factors such as temperature, pH, presence of other organic compounds etc. [8,9]. Many microorganisms have been reported for their ability to decolorize azo dyes such as bacteria, [10]. fungi, [11] actinomycetes [11] and algae. [12]. Researches showed that use of mixed culture can achieve a greater degradation of textile dye than a pure culture. [13].
Heavy metals are a special group of metals having a density greater than 5 g/cm3 that persist in nature and sequentially tend to accumulate in food chains. Though large amount of heavy metal present in the environment their presence in the food chain mainly due to the anthropogenic activities. [14]. The harmful effects of heavy metals on microorganisms depend on various abiotic factors such as concentration of chelating agents, speciation, pH and organic matter. [15,16]. Some heavy metals such as nickel, iron, copper and zinc serve as trace elements by the organisms and are essential for metabolic processes when others such as mercury, silver and cadmium have no biological role within the microbial cells and are harmful to the organisms, even at very low concentrations. [17]. Many bacteria have evolved genetic mechanisms to resist the toxic metals. [17]. These bacteria growing on environment heavily contaminated with high level of heavy metals can be used to remove heavy metal from soil and water by some modification of their degrading capability.
Various metals such as Mercury (Hg), Silver (Ag), Zinc (Zn), Copper (Cu), Chromium (Cr), Iron (Fe) and Mercury (Hg) were used for metal tolerance tests against the selected isolates. All the Isolates were isolated from soil and water from 8 distinct dye industry related places in Narayanganj District of Bangladesh. All the date of dye degrading capability and metals tolerance of the selected isolates were analyzed to find out the efficiency of a single bacterial isolate to degrade textile dye and remove heavy metal from soil and water that will be more economic and effective.
Materials and Methods
Sample collection
8 distinct dye industry related places in Narayanganj District of Bangladesh were primarily selected for samples collection.
Chemicals source
Different synthetic dyes were collected from Krishna kali Dyeing House of Gopaldi Bazar Pourashova, Araihazar, Narayanganj, Bangladesh.
Sample collection procedure
Samples were collected using distinct sterile Plastic Bottle and Plastic Polybag. Each Sample was collected from 8 distinct dye industries, transferred them aseptically to an ice box which maintained the temperature at 4ᴼC and then in microbiology laboratories at Noakhali Science and Technology University. Eight discrete samples were prepared for the study.
Physio-chemical analysis of textile effluents
The collected effluent samples have been analyzed to determine its Physical parameters. The various parameters such as pH, Color, Odor, Total solids (TS), Total dissolved solid (TDS), Total suspended solids (TSS), and Dissolved Oxygen (DO) were analyzed as described in standard methods for water and effluents analysis.
Biological analysis of textile effluents
Isolation of pure colony and preparation of pure culture: In this study, 10-1 to 10-3 dilutions of the different samples were used for preliminary isolation of dye decolorizing organisms. In order to isolate pure colony for this experiment, collected eight samples were directly plated on nutrient agar plate in the absence of synthetic dye and incubated at 37ºC for 24-72 hours. Each of plates was observed separately to find any kind of growth on the media, after incubation period. The distinct colonies were randomly isolated and streaked on nutrient agar plate to purify. Pure culture of 16 different isolates were prepared and stored.
Cultural and biochemical tests
Identification of bacteria: For initial screening of dye degrading bacteria, a loop of each sample was inoculated onto MacConkey agar media, Eosine Methylene Blue (EMB), Mannitol Salt Agar (MSA), Bacillus cereus Agar Base (BCAM), Cetrimide Agar media and incubated 37ºC for 24 hours. After the confirmation from cultural methods, all positive isolates of selected bacteria were subjected to different biochemical tests such as Gram staining, Shape, Oxidase, Catalase, TSI (Slant/Butt), Citrate test, VP test, MR test, Motility, Urease test, Indole test according to Bergeyes’s Manual of Bacteriology (NCCLS).
Stock culture preparation: After isolation of concerned organisms, organisms were cultured on nutrient agar plate.
After overnight incubation at 37ºC, each kind of organisms were picked and inoculated into TSB broth and stocked with 85% glycerol into an Eppendorf.
Dye decolorization assay: 100 ml Nutrient broth containing 0.12 gm/L different dyes were autoclaved at 121°C at 15 lbs pressure for 15 minutes. The flasks were inoculated with 2 ml of 24 hours old bacterial culture and incubated at 37°C up to 6 days. After centrifugation the cell free supernatants and cell containing pellets were used to determine the percentage decolorization of different selected dye at distinct wavelength by using spectrophotometer. The uninoculated dye medium supplemented with respective dye was used as control. Decolorization activity (%) was calculated by the following formula-
Decolorization (%) = [(Initial OD – Final OD)/Initial OD] × 100
Determination of heavy metal tolerance pattern
Heavy metal solution preparation: Heavy metals’ solution were preparation by dissolving the salt of each metal in distilled water up to make a solution of 1mg/ml concentration of the metals and sonicated for homogenous dissolving.
Determination of MIC of heavy metals: MIC of heavy metals was determined by serial dilution method. Serial dilution of the metals in the nutrient broth were prepared in such a way where the metals, salt concentration in the test tubes remained 10μg/ ml, 20 μg/ml, 30 μg/ml, 40 μg/ml, 50 μg/ml, 100 μg/ml, 200 μg/ ml, 300 μg/ml, 400 μg/ml and 500 μg/ml initially. Each type of bacterial species was used to determine their MIC against the heavy metals.
Results
Physical parameter of textile dye effluent samples
The collected sample have been analyzed to determine the some physicochemical characteristics like as color, pH, temperature, color, odor, TDS, TSS, TS, OD of industrial effluents. Table 1 indicates that the effluent samples color was varied greatly depending upon their collection point that may be due to the variation of dye and other residual compounds’ concentrations in the samples.
| S. No | Textile Dye Effluent Samples | Physico-chemical parameters | |||||||
|---|---|---|---|---|---|---|---|---|---|
| pH | Temperature °C | Color | odor | Total Dissolved Solids (in ppm) (g/L) | Total Suspended Solids (in ppm) (g/L) | Total Solids (in ppm) (g/L) | Dissolved Oxygen (mg/L) | ||
| 1 | S1 | 7.55 | 27.5 | Black-Grey | Unpleasant | 21400 | 11700 | 33100 | 37 |
| 2 | S2 | 7.63 | 27.4 | Red | Unpleasant | 15700 | 13500 | 29200 | 52 |
| 3 | S3 | 7.36 | 27.8 | Red | Unpleasant | 17400 | 16600 | 34000 | 29 |
| 4 | S4 | 6.89 | 27.5 | Red | Unpleasant | 14200 | 21400 | 35600 | 19 |
| 5 | S6 | 7.75 | 27.5 | Greenish | Unpleasant | 12700 | 19900 | 32600 | 53 |
| 6 | S7 | 7.34 | 27.4 | Red | Unpleasant | 14900 | 17600 | 32500 | 41 |
| 7 | B1 | 7.48 | 27.9 | Red | Unpleasant | 4200 | 8500 | 12700 | 34 |
| 8 | B3 | 7.87 | 27.5 | Greenish | Unpleasant | 3900 | 7800 | 11700 | 51 |
| 9 | B4 | 7.63 | 27.6 | Red | Unpleasant | 5400 | 13500 | 18900 | 27 |
Table 1: Physico-chemical parameters of textile dye effluent samples.
Identification and characterization of bacteria isolated from textile dye effluent
The identification and characterization of all isolates are performed by Bergey’s Manual of Systematic Bacteriology. In this present study twelve different bacterial isolates were isolated from eight dye effluent samples. The characteristics of the identified bacterial isolates were furnished in Tables 2 and 3. Based on morphological and biochemical characteristics, twelve bacterial isolates were identified as Staphylococcus spp., Bacillus spp., Pseudomonas spp., Enterobacter spp., Micrococcus spp. and Klebsiella spp. To carry on the study one isolate of each bacterial species was selected.
| Isolates Name | MacConkey agar media | Eosine Methylene Blue (EMB) | Mannitol Salt Agar (MSA) | Bacillus cereus Agar Base (BCAM) | Cetrimide Agar | Comments |
|---|---|---|---|---|---|---|
| Isolate1 | No growth | No growth | Yellow colony | No growth | No growth | Staphylococcus spp. |
| Isolate 2 | No growth | No growth | No growth | Blue-Yellow Colony | No growth | Bacillus spp. |
| Isolate 3 | No growth | No growth | No growth | Blue-Yellow Colony | No growth | Bacillus spp. |
| Isolate 4 | White or colorless colony | Pinkish mucoid colonies | No growth | No growth | Yellow – green to blue colonies | Pseudomonas spp. |
| Isolate 5 | Mucoid Central Red-Pink Colony | Light Violet | No growth | No growth | No growth | Enterobacter spp. |
| Isolate 6 | Mucoid Central Red-Pink Colony | Light Violet | No growth | No growth | No growth | Enterobacter spp. |
| Isolate 7 | No growth | No growth | Red Colony | No growth | No growth | Micrococcus spp. |
| Isolate 8 | Mucoid Central Red-Pink Colony | Light Violet | No growth | No growth | No growth | Enterobacter spp. |
| Isolate 9 | No growth | No growth | Red Colony | No growth | No growth | Micrococcus spp. |
| Isolate 10 | No growth | No growth | Yellow colony | No growth | No growth | Staphylococcus spp. |
| Isolate 11 | No growth | No growth | Yellow colony | No growth | No growth | Staphylococcus spp. |
| Isolate 12 | Pink Colony | Pale pink Colony | No growth | No growth | Yellow – green to blue colonies | Klebsiella spp. |
Table 2: Summary of result of cultural test on different isolates.
| Isolate Name | Gram staining | Shape | Oxidase | Catalase | TSI (Slant/Butt) | Citrate test | VP test | MR test | Motility | Urease test | Indole test | Comments |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate1 | + ve | Cocci | + ve | + ve | A/A | - ve | + ve | + ve | NM | - ve | - ve | Staphylococcus spp. |
| Isolate 2 | + ve | Rods | + ve | + ve | A/A | +ve | +ve | - ve | M | - ve | - ve | Bacillus spp. |
| Isolate 3 | + ve | Rods | + ve | + ve | K/A | +ve | +ve | - ve | M | - ve | - ve | Bacillus spp. |
| Isolate 4 | - ve | Rods | + ve | + ve | K/K | +ve | -ve | - ve | M | - ve | - ve | Pseudomonas spp. |
| Isolate 5 | - ve | Rods | - ve | + ve | A/A | - ve | -ve | + ve | M | - ve | + ve | Enterobacter spp. |
| Isolate 6 | - ve | Rods | - ve | + ve | A/A | - ve | -ve | + ve | M | - ve | + ve | Enterobacter spp. |
| Isolate 7 | + ve | Cocci | + ve | + ve | A/A | +ve | -ve | - ve | NM | + ve | - ve | Micrococcus spp. |
| Isolate 8 | - ve | Rods | + ve | + ve | K/A | - ve | -ve | + ve | M | - ve | + ve | Enterobacter spp. |
| Isolate 9 | + ve | Cocci | + ve | + ve | K/A | +ve | -ve | - ve | NM | + ve | - ve | Micrococcus spp. |
| Isolate 10 | + ve | Cocci | + ve | + ve | A/A | - ve | + ve | + ve | NM | - ve | - ve | Staphylococcus spp. |
| Isolate 11 | + ve | Cocci | - ve | + ve | A/A | - ve | +ve | + ve | NM | - ve | - ve | Staphylococcus spp. |
| Isolate 12 | - ve | Short rods | + ve | + ve | A/A | +ve | +ve | - ve | NM | + ve | - ve | Klebsiella spp. |
Table 3: Summary of result exemplified by biochemical analysis on different isolates.
Decolorization of textile dyes
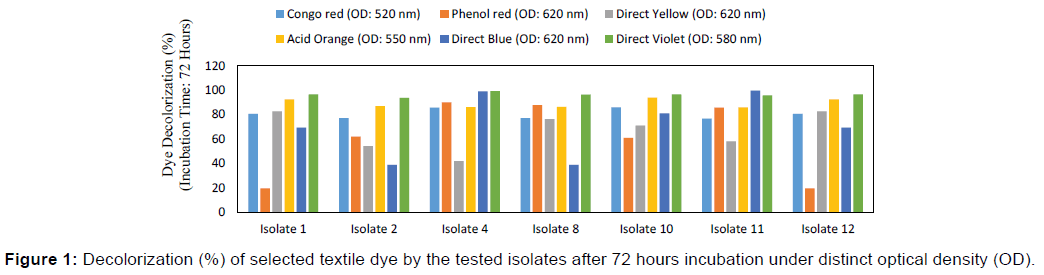
The percentage of dye decolorization was measured by spectrophotometer at different wavelengths. Six potential isolates namely Isolate 1, Isolate 2, Isolate 4, Isolate 8, Isolate 9 and Isolate 12 were tested for their ability to decolorize distinct selective local dyes namely Congo red, Phenol Red, Direct Yellow, Direct Red, Acid Orange, Direct Violet, Direct Blue and Direct Pink. Figure 1 provided the information about decolorization of selected distinct dye by the isolated bacteria. After 72 hours incubation, maximum decolorization of Congo red was up to 85.95% at the wavelength of 520 nm whereas Phenol red, Direct Blue and Direct Yellow were decolorized up to 90.03%, 99.64% and 82.68% at the wavelength of 620 nm respectively. Acid Orange was decolorized up to 93.96% at the wavelength of 550 nm and Direct Violet was decolorized up to 99.19% at the wavelength of 580 nm by six selected bacterial isolates.
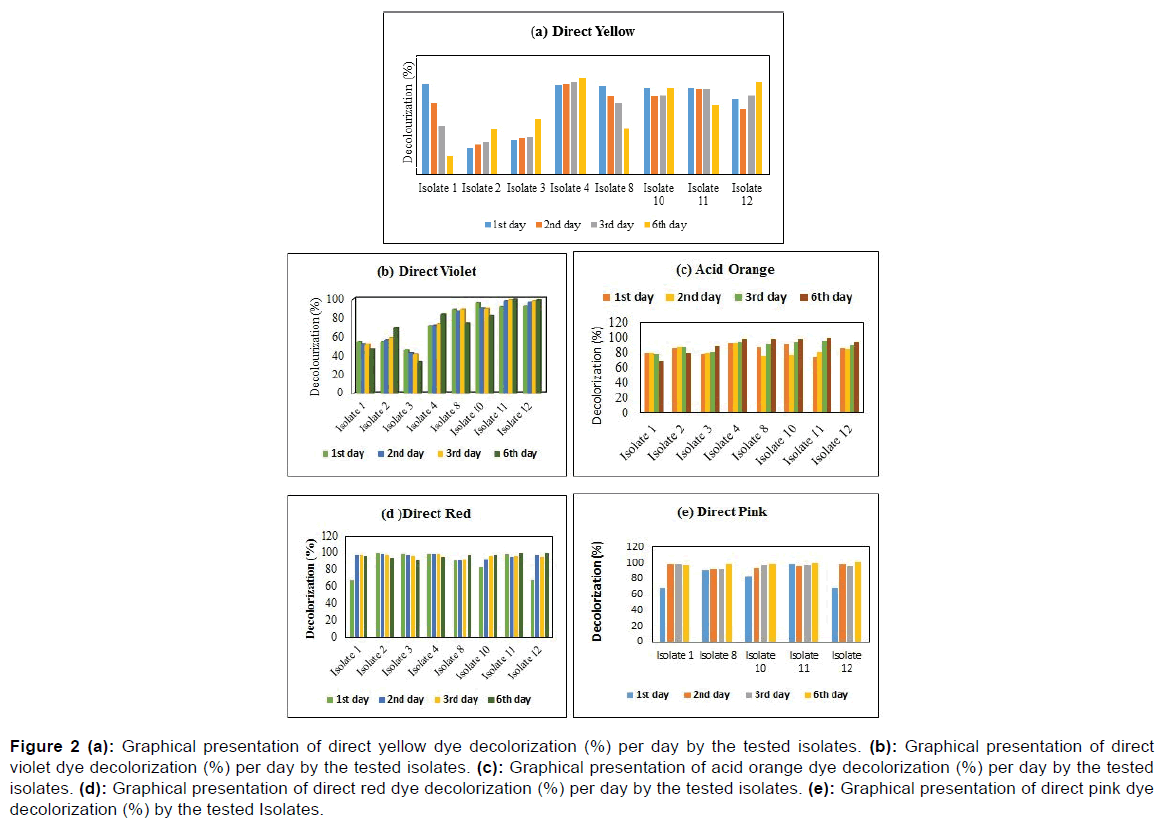
Decolorization (% per day) of selected textile under specific optical density (od 488nm) by spectrophotometer
The maximum percentage of decolorization of the Direct Yellow (488 nm) was about 90.32% (Isolate 1) whereas the lowest decolorization was 26.35% (Isolate 2) at 1st day while at the sixth day, Isolate 4 showed the maximum decolorization was about 96.98% while Isolate 1 revealed the minimum decolorization was recorded about 19.05%. Isolate 9 (96.11%) showed maximum decolorization activity of Direct Violet dye where the minimum one was recorded about 55.06% with Isolate 2 in the first day. However, 99.05% with Isolate 12 was maximum and 46.85% with Isolate 1 was minimum decolorization activity against respective dye for the time of last day.
Figure 2 showed that, Isolate 4 was the highest decolorization activity (93.21%) against Acid Orange dye (488 nm), the lowest decolorization activity was about 79.74% with Isolate 11 at the initial period. Then, in the sixth day, 98.78% with Isolate 9 was maximum as well as 67.68% with Isolate 1 was minimum decolorization activity against respective dye. As shown in Figure 2d, the local dye of direct red (488nm) was decolorized up to 67.74%, 99.64%, 99.10%, 99.10%, 90.91%, 82.89%, 98.40% and 67.74% respectively by Isolate 1, Isolate 2, Isolate 4, Isolate 8, Isolate 9 and Isolate 12 at the initial day. Then, at the sixth day 96.43%, 94.47%, 95%, 98.04%, 97.86% and 99.82% were accounted.
Figure 2: (a): Graphical presentation of direct yellow dye decolorization (%) per day by the tested isolates. (b): Graphical presentation of direct violet dye decolorization (%) per day by the tested isolates. (c): Graphical presentation of acid orange dye decolorization (%) per day by the tested isolates. (d): Graphical presentation of direct red dye decolorization (%) per day by the tested isolates. (e): Graphical presentation of direct pink dye decolorization (%) by the tested Isolates.
Figure 2e showed that, Isolate 4 was the highest decolorization activity (98.40%) against direct pink dye (488 nm), the lowest decolorization activity was about 67.74% with Isolate 1 and Isolate 12 at the initial period. At the 6th day Isolate 12 showed the highest decolorization activity while Isolate 1 and Isolate 2 showed the lowest decolorization activity against respective dye. The whole decolorization assay was done at the wavelength of 488 nm.
Minimum inhibitory concentration determination against different heavy metals
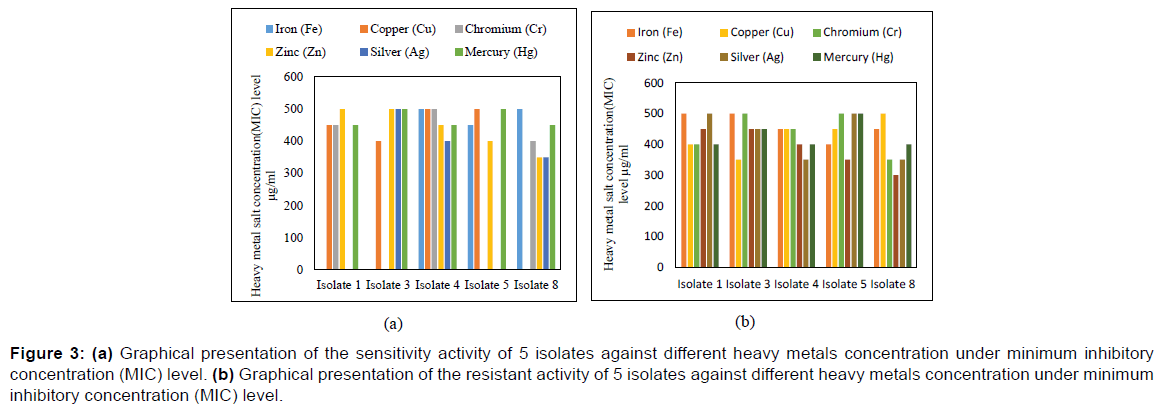
Minimum Inhibitory Concentration refers to the minimum concentration of at which bacterial growth can be inhibited. The growth pattern of selected isolates were determined ranging from 10 μg/ml to 550 μg/ml summarized on Figure 3 that represented the tolerance pattern of 6 isolates against different heavy metals concentration under Minimum Inhibitory Concentration (MIC) level.
Figure 3: (a) Graphical presentation of the sensitivity activity of 5 isolates against different heavy metals concentration under minimum inhibitory concentration (MIC) level. (b) Graphical presentation of the resistant activity of 5 isolates against different heavy metals concentration under minimum inhibitory concentration (MIC) level.
As shown in Figure 3a, Isolate 1, Isolate 2, Isolate 4, Isolate 8, Isolate 9 and Isolate 12 developed resistant activity against heavy metal Iron (Fe) under MIC level of 450 μg/ml, 550 μg/ml, 500 μg/ml, 500 μg/ml, 450 μg/ml and 400 μg/ml respectively within 24 hours where Isolate 2 showed maximum resistant activity under MIC level of 550 μg/ml. In case of Copper (Cu) Isolate 1, Isolate 2, Isolate 4, Isolate 8, Isolate 9, and Isolate 12 developed resistant activity under MIC level of 450 μg/ml, 400 μg/ml, 500 μg/ml, 550 μg/ml, 500 μg/ml and 450 μg/ml respectively within 24 hours where Isolate 8 showed maximum resistant activity under MIC level of 550 μg/ml. According to Figure 3, all the Isolate developed resistant activity against heavy metal Chromium (Cr) under MIC level of 450 μg/ml, 550 μg/ml, 500 μg/ml, 400 μg/ml, 550 μg/ml and 450 μg/ml respectively within 24 hours where Isolate 2 and Isolate 9 showed maximum resistant activity under MIC level of 550 μg/ml.
Isolate 1, Isolate 2, Isolate 4, Isolate 8, Isolate 9 and Isolate 12 developed resistant activity against heavy metal Zinc (Zn) under MIC level of 500 μg/ml, 500 μg/ml, 450 μg/ml, 350 μg/ml, 400 μg/ml and 350 μg/ml respectively within 24 hours. Here, Isolate 1 and Isolate 2 showed maximum resistant activity under MIC level of 500 μg/ml among them and all the isolates developed resistant activity against heavy metal Silver (Ag) under MIC level of 550 μg/ml, 500 μg/ml, 400 μg/ml, 400 μg/ml, 550 μg/ml and 450 μg/ ml accordingly where Isolate 1 and Isolate 9 showed maximum resistant activity under MIC level of 550 μg/ml.
As shown in Figure 3b, Isolate 1, Isolate 2, Isolate 4, Isolate 8, Isolate 9 and Isolate 12 developed resistant activity against heavy metal Mercury (Hg) under MIC level of 450 μg/ml, 500 μg/ml, 450 μg/ml, 450 μg/ml, 500 μg/ml and 400 μg/ml respectively within 24 hours where Isolate 2 and 9 showed maximum resistant activity under MIC level of 500 μg/ml. Among all the Isolates Bacillus spp., Micrococcus spp., and Pseudomonas spp. were found the most efficient heavy metal remover.
Discussion
Till now scientists are trying to find out an effective eco-friendly way to treat the textile dye but it is still a very big challenge. [18]. There are three types of methods for the treatment of textile dye namely physical, chemical and biological that have been extensively reviewed. [19,20]. Among the three methods of treatment biological treatment is more reliable which results, less sludge and more eco-friendly treatment. [21]. Various bacterial species have been reported to be capable of color removal and have the ability to decolorize diverse dyes efficiently compared to other. [21,22].
In this study, six isolates out of 12 isolates were screened for decolorization of different dyes. The used local dyes in this study belong to the metal azo dye group. They were selected for this study because of their wide and common use in the cotton textile industry in Bangladesh and worldwide. The biodecolorization of azo dyes vary depending on the presence of very specific changes in their molecular structures, [23]. and the different mechanisms of decolorization of the dyes which followed by different bacterial groups. [24]. Keeping all these in views, the present study was undertaken with the main aim of determination of Physio-chemical parameters, isolation and identification of dye decolorization bacteria, the decolorization percentage of textile azo dye with the help of spectrophotometer against distinct isolates and analyzing the resistance activity of tested bacteria under MIC level against respective metals.
The ability of microorganisms to remove color of single dyes efficiently does not necessarily mean the suitability of this organism in treatment of colored textile effluents. [25]. In nature the degradation of textile dye may be influenced by the wide range of pH, the high salt concentrations, the high temperature of effluent and the presence of chemical structures of additives. [18,25]. In this study, it was noticed that the percentage of dye decolorization using isolated bacteria was fluctuated at different wavelength based on different incubation periods. Based on 72 hours incubation period, minimum and maximum dye decolorization efficiency was found with isolated bacteria at discrete wavelength. This study reported that Isolate 9 (Micrococcus spp.) with Congo red and Acid Orange, Isolate 4 (Pseudomonas spp.) with Phenol red, Direct Blue and Direct Violet, Isolate 1 (Staphylococcus spp.) and 12 (Klebsiella spp.) with Direct Yellow were shown maximum decolorization ability of selected textile dyes. While, minimum dye decolorization efficiency was also observed with Isolate 2 (Bacillus spp.) and 8 (Enterobacter spp.) with Congo red and Direct Blue, Isolate 1 (Staphylococcus spp.) and 12 (Klebsiella spp.) with Phenol red, Isolate 4 (Pseudomonas spp.) with Direct Yellowand Acid Orange and isolate 2 (Bacillus spp.) with direct violet at the same period. This study was also reported that the Isolate 4 (Pseudomonas spp.) showed the most efficient decolorization of all selected textile dyes than others (Staphylococcus spp., Bacillus spp., Enterobacter spp., and Klebsiella spp.). Pseudomonas spp. was found to be more efficient in dye decolorization and facile conditions shows the potential for this bacterial strain to be used in the biological treatment of dyeing mill effluents. All the isolates showed resourceful tolerance against all the heavy metals used. Hence, this species can be used as a bioremediation tool for the treatment of effluent from leather and other industries handling heavy metals. Generally, exposure to a heavy metal increases the level of resistance of the bacterial community toward that metal. In the present study, it was found that the MIC of the selected heavy metals’ salts ranged from 350-550 μg/ml where Bacillus spp., Micrococcus spp., and Pseudomonas spp. were found the most efficient heavy metal remover. In another study, different bacterial isolates were found to tolerate various concentrations of heavy metals [25]. where the MIC for heavy metals varied between 5-3200 μg/ml. So by comparing the dye degrading capability and multi-metal resistant pattern of the selected bacterial isolates Pseudomonas spp. was found the most efficient as textile dye degrader and heavy metal remover. In a future, the techniques of molecular biology and biochemistry coupled with the latest advances in genomics and proteomics might offer a wide range of possibilities for enhancing the performance of these bacterial treatments of azo dye-containing wastewater as well as heavy metal contaminated water. [26]. On the other hand, a super bug may be constructed by the combination of heavy metal resistant gene and textile dye degrading gene. Mixed culture of textile dye degrading and heavy metal resistant bacteria can also be used to improve the treatment process. However, as the knowledge base and the funding in this area of research increases, it is hoped that bacterial treatments will become the predominant solution to the problem of colored wastewater contaminated with heavy metals in the textile coloration industry.
Conclusion
It is clearly indicated that industrial wastes are responsible for the development of textile dye and heavy metal resistance along with the risk of human health and environment. The long term effect of pollutants has led to emergence of dye degrading and multi-metal resistant bacteria in the study areas. The use of microbial populations specifically adapted to degrade textile dye and high concentrations of heavy metals can be used by increasing their ability to remediate textile dye from water as well as heavy metal contaminated water and soils respectively to lessen their rate in the human food chain.
Acknowledgements
It is up to us to express our deep and sincere gratitude to all the family of department of microbiology, Noakhali Science and Technology University, who assisted us in conducting this research.
Conflict of Interest
All authors disclose that there was no conflict of interest.
REFERENCES
- Tan NC, Borger A, Slender P, Svitelskaya AV, Lettinga G, Field JA. Degradation of azo dye Mordant Yellow 10 in a sequential anaerobic and bioaugmented aerobic bioreactor. Water Sci. Tech. 2000; 42: 337-344.
- Carliell CM, Barclay SJ, Naidoo N, Buckley CA, Mulholland DA, Senior E. Microbial decolourization of a reactive azo dye under anaerobic conditions. Water SA. 1995; 21: 61-69.
- Peralta-Zamora P, Pereira CM, Tiburtius ERL. Decolorization of reactive dyes by immobilized laccase. Applied Catalysis B: Environmental. 2003; 42: 131-144.
- Rajaguru P, Vidya L, Baskarasethupathi B, Kumar PA, Palanivel M, Kalaiselvi K. Genotoxicity evaluation of polluted ground water in human peripheral blood lymphocytes using the comet assay. Mutat. Res. 2002; 517: 29-37.
- Umbuzeiro GA, Freeman H, Warren SH, Oliveira DP, Terao Y, Watanabe T, et al. The contribution of azo dyes to the mutagenic activity of the Cristais River. Chemosphere. 2005; 60: 55-64.
- Kunz A, Mansilla H, Durán N. A degradation and toxicity study of three textile reactive dyes by ozone. Environ. Tech. 2002; 23: 911-918.
- Wang H, Su JQ, Zheng XW, Tian Y, Xiong XJ, Zheng TL. Bacterial decolorization and degradation of the reactive dye Reactive Red 180 by Citrobacter sp. CK3. Int. Biodeterior. Biodegradation. 2009; 63: 395-399.
- Deng D, Guo J, Zeng G, Sun G. Decolorization of anthraquinone, triphenylmethane and azo dyes by a new isolated Bacillus cereus strain DC11. Int. Biodeterior. Biodegradation. 2008; 62: 263-269.
- Forgacs E, Cserháti T, Oros G. Removal of synthetic dyes from wastewaters: A review. Environ. Int. 2004; 30: 953-971.
- Carliell CM, Barclay SJ, Shaw C, Wheatley AD, Buckley CA. The effect of salts used in textile dyeing on microbial decolourisation of a reactive azo dye. Environ. Tech. 1998; 19: 1133-1137.
- Zheng Z, Levin RE, Pinkham JL, Shetty K. Decolorization of polymeric dyes by a novel Penicillium isolate. Proc. Biochem. 1999; 34: 31-37.
- Dilek FB, Taplamacioglu HM, Tarlan E. Colour and AOX removal from pulping effluents by algae. Appl. Microbiol. Biotechnol. 1999; 52: 585-591.
- Nigam P, Banat IM, Singh D, Marchant R. Microbial process for the decolorization of textile effluent containing azo, diazo and reactive dyes. Proc. Biochem. 1996; 31: 435-442.
- Trevors JT, Oddie KM, Belliveau BH. Metal resistance in bacteria. FEMS Microbiol. Rev. 1985; 32:39-54.
- Sterrit RM, Lester JN. Interactions of heavy metals with bacteria. Sci. Total Environ. 1980; 14: 5-17.
- Hughes MN, Poole RK. The functions of metals in micro-organisms. In: Hughes, M.N.; Poole, R.K. (eds). Metals and microorganisms. Chapman and Hall, London. 1989; p. 1-38.
- Silver S, Misra TK. Plasmid-mediated heavy metal resistances. Ann Rev Microbiol. 1988; 42: 717-743.
- Dos Santos AB, Cervantes FJ, Van Lier JB. Review paper on current technologies for decolourisation of textile wastewaters: perspective for anaerobic biotechnology. Bioresource Technol. 2007; 98: 2369-2385.
- Hao OJ, Kim H, Chiang PC (2000). Decolourization of wastewater. Crit Rev Environ Sci Technol. 30: 449-505.
- Robinson T, Chandran B, Nigam P. Studies on the decolorization of an artificial textile effluent by white-rot fungi in N-rich and N-limited media. Appl Microbiol Biotechnol. 2001; 57: 810-813.
- Saraswathy RN, Shanmugapriya S, Shakthipriyadarshini S, Sadasivam S, Shanmugaprakash M. Decolorization of textile dyes by Aspergillus tamarii, mixed fungal culture and Penicillium purpurogenum. Journal of Science and Industrial Research. 2010; 69: 151-153.
- Fu Y, Viraraghavan T. Fungal decolorization of dye wastewaters. A review. Bioresource Technol. 2001; 79: 251-262.
- Park C, Lee M, Lee B, Kim SW, Chase HA, Lee J, et al. Biodegradation and biosorption for decolorization of synthetic dyes by Funalia trogii. Biochem. Eng. J. 2007; 36: 59-65.
- Wilkolazka AJ, Rdest JK, Malarczyk E, Wardas W, Leonowicz A. Fungi and their ability to decolourize azo and anthraquinonic dyes. Enzyme Microb. Technol. 2002; 30: 566-572.
- Mester T, Tien M. Oxidation mechanism of ligninolytic enzymes involved in the degradation of environmental pollutants. Int. Biodeterior. Biodegradation. 2000; 46: 51-59.
- Matyar F, Kaya A, Dinçer S. Antibacterial agents and heavy metal resistance in Gram-negative bacteria isolated from seawater, shrimp and sediment in Iskenderun Bay, Turkey. Sci Total Environ. 2008; 407: 279-285.




 The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.
The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.