Investigation of 13q14.3 Deletion by Cytogenetic Analysis and FISH Technique and miRNA-15a and miRNA-16-1 by Real Time PCR in Chronic Lymphocytic Leukemia
2 Department of Medical Biology, Istanbul University, Cerrahpasa, Istanbul, Turkey, Email: dilhankuru@yahoo.com
3 Department of Hematology, Istanbul University, Cerrahpasa, Istanbul, Turkey
Citation: Yilmaz M, et al. Investigation of 13q14.3 Deletion by Cytogenetic Analysis and FISH Technique and miRNA-15a and miRNA-16-1 by Real Time PCR in Chronic Lymphocytic Leukemia. Ann Med Health Sci Res. 2021;11:1403-1410
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Background:The most frequent cytogenetic aberration is 13q14.3 deletion in Chronic Lymphocytic Leukemia (CLL). Hsa-miR-15a/hsa-miR-16-1 are tumor suppressor miRNAs encoded from 13q14.3 region. Objectives: The aim of this study was to investigate the 13q14.3 deletion using molecular and cytogenetic techniques and association with miRNA-15a/miRNA-16-1. Materials & Methods: We used peripheral blood samples of 30 CLL patients which were either induced and or non-induced with DSP30+IL-2 for determine 13q14.3 deletion by karyotyping and iFISH methods. Expression levels of hsa-miR-15a/miR-16-1 were measured using Quantitative Real Time PCR and compared with deletions. Results: 13q14.3 deletion was detected in 8.6% of cases by karyotyping and in 65% by iFISH. Mosaic forms (monoallelic+biallelic) were observed in 50% of cases. Besides determining common chromosome abnormalities such as add(2)(q37), t(2;7)(p11.2;q22), del(6)(q13q21), del(6)(q25), add(9)(q21), del(11) (q23), t(11;14)(q13;q32), del(13)(q11q12), del(13)(q12q14), add(14)(q23), del(14)(q23), t(14;19)(q32;q13.1), del(15)(q23), del(17)(p12), t(18;22)(q21;q11.2), add(21)(p13) and t(17;21)(q11.2;122), we also determined t(1;13)(q32;q34), inv(2)(p25q21), del(13) (q22q32), t(14;19)(q24;q13), dup(17)(q21q23), rob(21;21)(p13;p13) which have not been reported previously. Mitotic index data was found statistically significant and DSP30+IL-2 increased mitotic index by 2.5 folds. Association between decreased miR- 16-1 expression and deletions was statistically significant. Conclusion: We suggest that cytogenetic and iFISH analyses are complementary and use of DSP30+IL-2 is effective in CLL. Decreased expression of hsa-miR-16-1 is remarkable.
Keywords
CLL; miR-15a/miR-16-1; 13q14.3 deletion; DSP30; iFISH; qRT-PCR
Introduction
Chronic Lymphocytic Leukemia (CLL) is characterized by the gradual accumulation of functionally immature, small, monoclonal CD5+ and CD23+ B cells most of which are nonproliferating cells arrested at G0/G1 phase of the cell cycle. [1] CLL comprises about 30% of all cases of adult leukemia in Western World affecting both sexes and chromosomal abnormalities are widely used in CLL to prognose, treat and follow the overall survival in patients. [2] These abnormalities include del(13)(q14), del(11)(q22), trisomy 12 and del(17)(p13) and they can be detected using conventional banding techniques. The most frequent abnormality is deletion of 13q14 which is associated with favorable prognosis. [3,4] Deletion of 13q14 is mostly monoallelic (in 76% of cases), but also it is detected in biallelic (24%) and mosaic forms by iFISH. [4,5]
Conventional karyotyping and Interphase Fluorescence In Situ Hybridization (iFISH) techniques are widely used to detect cytogenetic abnormalities seen in CLL. However, approximately 50% of patients with CLL can be analyzed by conventional karyotyping due to very low responsiveness of CLL cells to mitogenic stimuli in vitro. [3] Recently a new mitogen CpGoligodinucleotide (CpD-ODN) called DSP30 was used in CLL cytogenetics which was reported to be the most effective CpGODN in stimulating human B cells. [6]
MicroRNAs (miRNAs) are 20–22 nucleotide long small noncoding RNAs which can bind to Untranslated Regions (UTRs) of target mRNAs resulting in translational repression or mRNA degradation. [7] DLEU2 gene encoding miR-15a and miR-16-1 are located within the deletion region of 13q14. Both miR-15a and miR-16-1 have been shown to exhibit tumorsuppressing activities by inducing apoptosis and inhibiting proliferation. Generally speaking, levels of both miRNAs are decreased due to deletion 13q14 but physical loss of 13q14 is not the only mechanism causing a decrease in miRNAs levels. Epigenetic regulations and defects in miRNA biogenesis can contribute to their dysregulation as well. [8,9] In this study, we investigated miR-15a and miR-16-1 levels by qRT-PCR and compared with 13q14.3 deletion detected by conventional karyotyping and iFISH techniques. We also investigated other possible cytogenetic abnormalities and effects of DSP30+IL-2 combination using conventional karyotyping techniques in patients with CLL.
Methods
Peripheral blood was collected from 30 CLL patients. All patients were followed up in division of hematology. 18 (60%) of patients are males and 12 (40%) are females. 15 healthy control subjects 9 (60%) males and 6 (40%) females) were also included in the study. Expression levels of miR-15a and miR- 16-1 were measured using qRT-PCR, while deletion of 13q14 region was assessed using conventional cytogenetic and iFISH techniques. Informed consent was obtained from mall patients prior to the study and all study was conducted following declaration of Helsinki. The Ethical Committee of Istanbul University Cerrahpasa Medical Faculty approved the study (nr:83045809/604/01-01/114304).
Conventional cytogenetic analysis using DSP30+IL-2
Conventional karyotyping analysis was performed in all patients using standard protocols. Peripheral blood samples (~1 ml) of patients were added in three separate conical tubes and suspended in 2.5 ml of 1640 RPMI (Biochrome, Berlin, Germany). We added 10 μl of CpG-ODN DSP30 (TıbMolBiol, Berlin, Germany) with a concentration of 1μM and 50 μl IL-2 (Roche, Mannheim, Germany) with a concentration of 100 U/ mL for both first and second tubes. Third tubes were used as control which does not include DSP30 and IL-2. All samples were cultured at 37℃ for 72 h. 50 μl of colcemid (KaryoMax, Thermo Fisher Scientific, Waltham, MA, USA) (with a final concentration of 10 μg/ml) was added prior to harvesting of the cells.
Chromosome preparation and staining using G-Banding technique was performed following standard procedures in patients and control samples, as described elsewhere. [10] Karyotypes were scored according to the International System for Human Cytogenetic Nomenclature (ISCN) 2016. [11] Mitotic index was scored by counting metaphases on a slide containing 1000 cells per mitosis in induced by DSP30 and noninduced patient samples and compared to the controls.
iFISH experiments
iFISH analyses were performed in interphase cells using 13q14.3 locus specific probe (Cytocell LSI-D13S319 Plus, Cambridge, UK) in induced samples by DSP30 of all patients. Cultured samples were re-suspended with fresh Carnoy fixative (3:1 methanol:acetic acid) for two hours at Room Temperature (RT), centrifuged, removed the supernatant, and then re-suspended in fresh fixative for a few more minutes before spreading and then CytocelliFISH protocol was performed. [12] Three hundred interphase nuclei were scored in all samples. Same protocols were applied for the control samples.
qRT-PCR experiments
miRNA was isolated using ExiqonmiRCURY RNA isolation kit (Exiqon A/S, Vedbaek, Denmark) following the manufacturer’s instructions and cDNA was synthesized using miRCURY LNA™ microRNA PCR, Polyadenylation and cDNA synthesis kit II (Exiqon A/S, Denmark) with an initial amount of 10 ng of RNA. cDNA synthesis was performed in T100 Thermal Cycler System (Bio-rad Laboratories, Hercules, CA, USA) with following conditions: 42°C for 60 min, 95°C for 5 min and 4°C for cooling. The expression levels of miR-15a and miR-16-1 were determined with a quantitative system based on SYBR Green probe technology [miRCURY LNA™ microRNA PCR, ExiLENT SYBR® Green master mix (Exiqon S/A, Denmark)]. The assay was performed in a total volume of 10 μl and contained 5 μl 1xPCR master mix, 1 μl of each primer
Statistical analyses
Statistical analyses were performed with the SPSS 21 software (IBM Corp. Released 2012. IBM SPSS Statistics for Windows, Version 21.0. Armonk, NY: IBM Corp). Wilcoxon test was used to confirmation of mitotic index in DSP30+IL-2 induced and non-induced samples and mitotic index differences between patient and control samples (p<0.001). Cut-off values of deletion types in patients and control samples were calculated using ROC analysis. Association between miRNAs (hsa-miR- 15a, hsa-miR-16-1) expression levels and 13q14.3 deletion types were analyzed using Mann-Whitney U test. Significance value was determined as (p<0.05).
Results
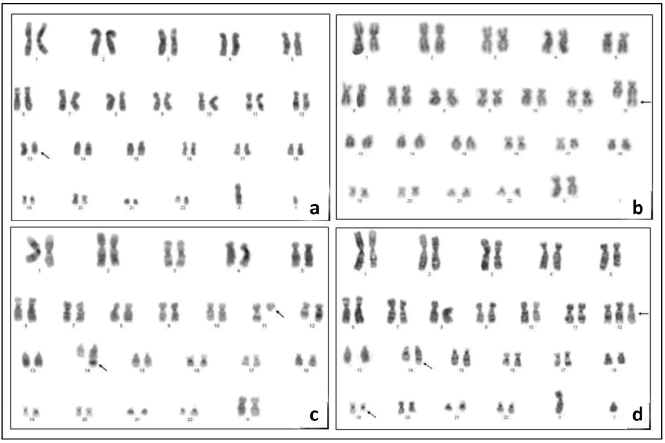
In DSP30+IL-2 induced samples, the sufficient number of metaphases suitable for cytogenetics analysis was obtained in 23/30 (76.6%) of the patients and clonal chromosomal aberration were detected in 16/23 (69.5%) of the patients. Normal karyotype was found in 7/23 (30.4%) of the patients. 13q14.3 deletion was detected only in 2/23 (8.6%) of the patients with conventional karyotyping techniques. In DSP30+IL-2 noninduced samples metaphases suitable for cytogenetics analysis were observed in 7/30 (23.3%) of the patients and chromosomal aberrations were detected in 2/7 (28.5%) of the patients. 13q14.3 deletions was not observed in non-induced samples by conventional karyotyping. Cytogenetic aberrations of twentythree successfully karyotyped patients are presented in Table 1. Also some different abnormalities we found in induced samples are shown in Figure 1.
| Caseno. | DSP30+IL-2 induced | DSP30+IL-2 non-induced |
|---|---|---|
| 1 | - | - |
| 2 | - | - |
| 3 | 46, XY(22) | - |
| 4 | 46, XX(14) | - |
| 5 | 41~46,XY, -Y(3),t(2;7)(p11.2;q22)(28),del(6)(q?q?) (9), 9qh+c(19),der(17)t(3;17)(q21;p11.2)(29),-21(5),+21(5),add(21)(p13)(6),rob(21;21)(p13;p13)(14),mar1(3)(cp31),/46,XY,9qh+c(2) | - |
| 6 | 46, XY,del(17)(p12),dup(17)(q21q23)(3)/46~47,XY,+13,del(13)(q11q12)(cp2) /46,XY(4) | 46, XY(4) |
| 7 | 46,XX,der(13)(q12q14)del(3)(q22q32)(8)/38~46,XX,del(6)(q?q?) (12),der(13)del(13)(q12q14) del(13)(q22q32)(16),-17(4),t(17;21)(q11.2;q22)(2),der(21)t(17;21)(q11.2;q22)(2),-21(3),-22(3)(cp19) /46,XX(2) | - |
| 8 | 40~45, XY, -20(cp4)/46, XY(19) | 46, XY(3) |
| 9 | 46, XY(8) | - |
| 10 | - | - |
| 11 | 43~49,XY,+8(1),del(14)(q23)(21)(cp21)/92~98,XXY,+4(2) +8(1),+14(2) del(14)(q23)x2(4),+19(2)(cp4) /46,XY(2) | 45, XY, del(14)(q23)(cp1)/ 46, XY(1) |
| 12 | 46,XY,t(1;13)(q32;q34)(3),del(6)(q13q21)(2)(cp3)/ 46,XY(8) | - |
| 13 | 46,XY(19) | 46, XY(11) |
| 14 | - | - |
| 15 | 42~46,XX,-10(3),t(11;14)(q13;q32)(10),-21(3)(cp12) /46,XX(9) | - |
| 16 | 46,XX,del(11)(q23)(2)/46,XX(5) | - |
| 17 | 39~46, XY, t(18;22)(q21;q11.2)(11), -21(3)(cp11) /46, XY(5) | - |
| 18 | - | - |
| 19 | 47, XY, +12(22), add(14)(q32)(3), t(14;19)(q32;q13.1) (15),t(14;19)(q24;q13)(2)(cp22) | 43~48, XY, +12(10), t(14;19)(q32;q13.1)(4),add(14)(q32)(3),t(14;19)(q24;q13)(1)(cp10) |
| 20 | 41~46, XY, +12(20), -21(3)(cp21)/46, XY(2) | - |
| 21 | 46, XX, dup(12)(q13q24)(2)/46, XX(24) | - |
| 22 | 46, XX, add(2)(q37), del(6)(q25), del(15)(q23), +mar1(2)/ 46, XX, inv(2)(p25q21)(cp2)/46, XX(40) | 46, XY(1) |
| 23 | 46, XX(10) | - |
| 24 | 42~45, XX, -21(cp4)/46, XX(16) | - |
| 25 | 46, XY(16) | - |
| 26 | 46, XX, +20(cp2)/46, XX(8) | - |
| 27 | 46, XX(2) | - |
| 28 | - | - |
| 29 | - | - |
| 30 | 46, XY, del(13)(q12q14)(10)/46, XY(7) | - |
(-): No results are available due to absent cells and/or low mitotic index
Table 1: Conventional cytogenetic results in samples induced and non-induced with DSP30+IL-2.
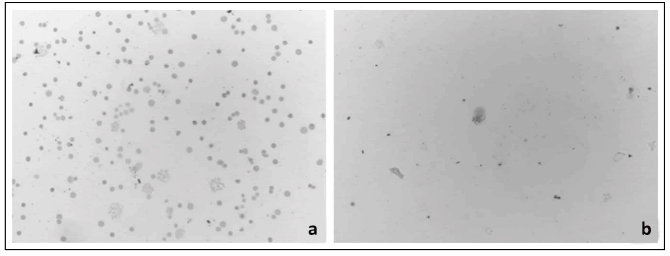
Mitotic index was calculated as 10.48% in DSP30+IL-2 induced samples and as 4.2% in non-induced samples whereas defıned as 45% in control subjects. We identified that using DSP30 increased mitotic index by 2.5 folds. Difference in induced and non-induced samples was found statistically significant (p<0.001) [Table 2]. Metaphase plaques (number of metaphases) and cell density in induced and non-induced samples are shown in Figure 2.
| Induced with DSP30+IL-2 | Non-induced with DSP30+IL-2 | P value | |
|---|---|---|---|
| Patient Median (min-max) | 3.5 (0-46.8) | 0 (0-2.50) | <0.001 |
Table 2: Results of Mann-Whitney Test Statistic used for compare of mitotic index with using and non-using DSP30+IL-2 samples.
13q14.3 deletion was detected in 2/23 (8.6%) of DSP30+IL-2 induced patients by conventional karyotyping whereas this deletion was detected by iFISH in 17/26 (65.3%) of the same patients. Monoallelic and mosaic deletions were determined in 17/26 (65.3%) and in 13/26 (50%) of the patients, respectively. Statistically; cut-off values, performed by ROC analyses (p<0.05) [Table 3], were calculated as 1.88% and 0.13% for monoallelic (2G/1R) and biallelic (2G) deletions, respectively.
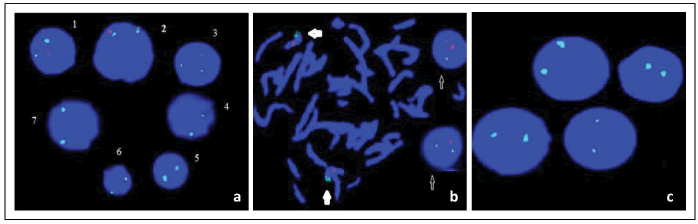
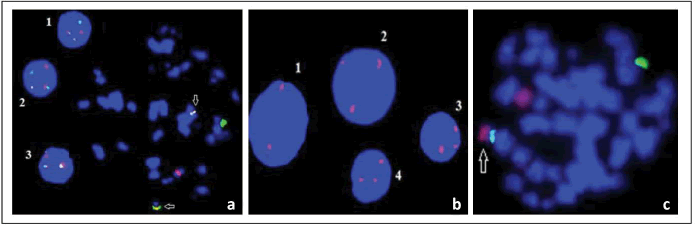
iFISH images of mono and biallelic deletions at 13q14 and other chromosomal abnormalities except 13q14 that were detected in DSP30+IL-2 induced samples are shown in Figure 3 and Figure 4, respectively.
| Test result variable(s) (%) | Area | Std. Error | p value | 95% Confidence interval |
|---|---|---|---|---|
| 2G/2R (Normal) | 0.314 | 0.077 | 0.014 | 0.164-0.465 |
| 2G/1R (Monoallelic D.) | 0.674 | 0.076 | 0.022 | 0.523-0.823 |
| 1G/1R (Monosomy) | 0.606 | 0.078 | 0.163 | 0.452-0.759 |
| 2G (Biallelic D.) | 0.737 | 0.069 | 0.002 | 0.601-0.873 |
Table 3: Results of ROC analysis for del(13q14.3).
Figure 3:FISH images of monoallelic and biallelic deletions of 13q14.3 in different patient samples induced with DSP30+IL-2: (a) iFISH images of monoallelic (1, 2, 3) and biallelic (4, 5, 6, 7) 13q14.3 deletions, (b) Metaphase FISH (mFISH) images of normal cell and iFISH images of monoallelic deletion cells, (c) iFISH images of biallelic 13q14.3 deletions cells (Green signal:Control=13qter/13q34/Red signal: Deletion=13q14).
Figure 4:FISH images of different chromosomal abnormalites in DSP30+IL-2 induced samples: (a) iFISH(1,2,3) and mFISH images of t(11;14 ) (q13.3;q32.33) (Red Signal,11q13.3; Green Signal,14q32.33), (Yellow Signal, Fusion (t(11;14)(q13.3;q32.33)), (b) iFISH images of normal (1) and trisomy 12 (2,3,4) cells (Red Signal,Centromere:12/12p11.1-q11.1/D12Z3), (c) mFISH image of der(21)t(17;21) (Red signal, chr.17; Green Signal, chr. 21, arrow:fusion chromosome).
We showed that miRNA-15a expression was decreased in 13/30 (45.5%), increased in 15/30 (50%), not changed in 2/30 (6.6%) of the patients whereas miR-16-1 expression decreased in 15/30 (50%), increased in 14/30 (46.6%), not changed in 1/30 (3.3%) of the patients when compared to controls. Expression levels of both miRNAs were simultaneously decreased in 7/30 (23.3%) and increased in 8/30 (26.6%) of the patients. Hsa-miR-15a and hsa-miR-16-1 expression changes are shown in Table 4. Statistical analyses showed that changes were not significantly correlated with deletions of 13q14 [Table 5] but decreased miRNA-16-1 expressions were found to be correlated [Table 6].
| Case no | miR-15a | miR-16-1 | Patient No | miR-15a | miR-16-1 |
|---|---|---|---|---|---|
| 1 | decreased | decreased | 16 | increased | decreased |
| 2 | increased | increased | 17 | increased | increased |
| 3 | increased | increased | 18 | increased | increased |
| 4 | decreased | not change | 19 | decreased | decreased |
| 5 | increased | decreased | 20 | increased | increased |
| 6 | decreased | decreased | 21 | increased | increased |
| 7 | decreased | decreased | 22 | increased | decreased |
| 8 | decreased | decreased | 23 | not change | decreased |
| 9 | decreased | increased | 24 | increased | decreased |
| 10 | decreased | decreased | 25 | increased | increased |
| 11 | decreased | increased | 26 | decreased | increased |
| 12 | increased | decreased | 27 | decreased | decreased |
| 13 | increased | increased | 28 | decreased | increased |
| 14 | increased | increased | 29 | decreased | increased |
| 15 | increased | decreased | 30 | increased | decreased |
Table 4: Expression patterns of miR-15a and miR-16-1 in patients.
| (hsa-miR-15a) | ||||
|---|---|---|---|---|
| 2G/2R | 2G/1R | 1G/1R | 2G | |
| P value | 0.913 | 0.477 | 0.701 | 0.542 |
Table 5: Mann-Whitney U test P values showing the association between the expression levels of hsa-miR-15a gene and 13q14.3 deletions.
| (hsa-miR-16-1) | ||||
|---|---|---|---|---|
| 2G/2R | 2G/1R | 1G/1R | 2G | |
| P value | 0.017 | 0.008 | 0.585 | 0.007 |
Table 6: Mann-Whitney U test P values showing association between expression levels of hsa-miR-16-1a gene and 13q14.3 deletions.
Discussion
CLL is a blood and bone marrow disease developing slowly over time resulting with over production of lymphocytes by bone marrow mostly affecting older adults. Many genetic and epigenetic factors contribute to the formation of the disease. [14- 16] Therefore, detecting molecular and cytogenetic abnormalities has vital importance in diagnosis and treatment of the disease. [17,18] However, it is extremely difficult to diagnose the abnormalities using only conventional karyotyping techniques due to the low mitotic index of lymphocytes. Stimulating the CLL cells using various agents for cytogenetic analysis has major importance in diagnosis. One of these effective immunostimulatory agents is DSP30. Recent studies have shown the efficacy of DSP30 on CLL cells either alone or with another agent (IL-2). [17-22]
In this study, we used conventional karyotyping, iFISH and qRT-PCR techniques in 30 CLL patients and 15 healthy controls to detect the abnormalities developed in CLL. As karyotyping is considered as a low efficient method due to low proliferation of CLL cells, iFISH technique and DSP30 have found a wider area of application to stimulate CLL lymphocytes in order to detect chromosomal abnormalities.
Regarding the chromosome banding, we found that clonal chromosomal abnormalities were present in 16/23 (69.5%) patient samples induced with DSP30+IL-2 using conventional karyotyping method. The occurrence of del(13q), del(11q), +12, and del(17p) was established in 8.6% (2/23), 4.3% (1/23), 8.6% (2/23) and 4.3% (1/23), respectively. These values are comparable to the data published so far.[18,23,24] Other chromosomal abnormalities we determined are shown in Table 1 and common abnormalities diagnosed in CLL causing bad prognosis such as t(11;14)(q13;q32), [25-27] dup(12)(q13q22), t(14;19)(q32;q13.1) accompanying with +12 andadd(14)(q32) [26-28] were also present in our study.
Trisomy 12 mechanism and target genes are not clear in CLL and associate with t(14;19)(q32;q13). So; previous studies and our results support that these three abnormalities can be clonal changes in leukemiagenesis of CLL and investigating this mechanism can be beneficial in order to understand the pathogenesis of CLL and develop new treatment options. We also determined t(1;13)(q32;q34), inv(2)(p25q21),t(3;17) (q21;p11.2),del(13)(q22q32), t(14;19)(q24;q13), dup(17) (q21q23), rob(21;21)(p13;p13) which have not been reported previously. To our knowledge, this is the first study reporting t(14;19)(q24;q13) in CLL. Previous studies have reported t(14;19)(q32;q13.1) in CLL. However, our new translocation spans a larger area on chromosome starting from q24 region. This large area may be associated with CLL. Balanced rearrangements are rare in CLL and we detected balanced translocation rob(21;21)(p13;p13).
In non-induced samples; we determined successful karyotyping in 6/30 (20%) of the patients and detected clonal chromosomal abnormalities in 2/6 (33%) of the patients. However, no chromosomal abnormalities were detected in 4 induced samples. Also we determined that DSP30+IL-2 caused an increase in mitotic index by 2.5-folds. So our results showed that using DSP30+IL-2 in CLL patients is very effective to increase mitotic index which allows us to detect sufficient number and quality metaphases and detect chromosomal abnormalities by inducing division of leukemic cells as reported in other studies. [17-20]
We investigated 13q14.3 deletion with both conventional karyotyping and iFISH techniques. del(13q) was detected only in 8% of patients with karyotyping while iFISH revealed it in 65.3%. Previous studies have reported rates varying between 1%-56%. [18,23,29,30] Similar to previous studies, our results indicate that iFISH technique is a very sensitive and specific method for routine diagnosis of CLL. In order to detect the abnormalities seen in chromosomes, conventional karyotyping is the most common method. On the other hand, it is obligatory to know the abnormality in advance to apply iFISH method which uses specially designed probes. Use of both methods at the same time can yield better results. In conclusion, DSP30+IL-2 should be preferred for routine conventional karyotyping of CLL and both techniques should be complementarily applied.
Our study showed decreased expression of miR-16-1 which correlates with monoallelic and biallelic deletion. Recent studies reported decreased expression of miR-15a and miR-16- 1 in CLL cases. Calin et al. reported decreased miR expression in their study. [16] Smonskey et al. showed that the decrease in miR-15a expression correlated with deletions but no significant association between miR-16-1 expression and deletions was found in their study. [9]
Conclusion
In conclusion, we didn’t find any correlation between miR-15a expression levels and deletions but some patients with del(13q) had increased miRNA expression levels or some patients without del(13q) decreased expression levels. miR-16-1 expression was found to be decreased but it was not related to deletions. So these findings can suppose that expression of both miRNAs not only regulates chromosomal loss but also other mechanisms such as epigenetic changes. Also, increased expression levels of both miRs in del(13q) positive patients may be due to the presence of homologous gene loci miR-15b/miR-16-2 in the 3q25-26.1. Our results indicate that miR16-1 plays an effective role in CLL pathogenesis when compared with miR-15a.
According to our results, FISH method is more specific to detect deletion of 13q14.3 than conventional karyotyping methods. We also suggest that using DSP30+IL-2 combination can be very useful for karyotyping methods of CLL. Also miR-16-1 expression levels can be used to investigate in CLL patients with del(13q).
Competing Interests
The authors of this paper have no conflicts of interest, including specific financial interests, relationships, and/or affiliations relevant to the subject matter or materials included.
REFERENCES
- Sagatys EM, Zhang L. Clinical and laboratory prognostic indicators in chronic lymphocytic leukemia. Cancer Control. 2012;19:18-25.
- Chena C, Avalos JS, Bezares RF, Arrossagaray G, Turdó K, Bistmans A, et al. Biallelic deletion 13q14. 3 in patients with chronic lymphocytic leukemia: cytogenetic, FISH and clinical studies. Eur J Haematol. 2008;81:94-99.
- Vroblová V, Smolej L, Vrbacky F, Jankovicová K, Hrudková M, Maly J, et al. Biological prognostic markers in chronic lymphocytic leukemia. ActaMedica. 2009;52:3-8.
- Put N, Wlodarska I, Vandenberghe P, Michaux L. Genetics of chronic lymphocytic leukemia: practical aspects and prognostic significance. Chronic Lympho Leuk. 2012:269.
- Humplikova L, Kollinerova S, Papajik T, Pikalova Z, Holzerova M, Prochazka V, et al. Expression of miR-15a and miR-16-1 in patients with chronic lymphocytic leukemia. Biomed J. 2013;157:284-293.
- Hartmann G, Krieg AM. Mechanism and function of a newly identified CpG DNA motif in human primary B cells. J Immunol.2000;164: 944-953
- Undi RB, Kandi R, Gutti RK. MicroRNAs as haematopoiesis regulators. Adv Hematol. 2013;1-20.
- Deng S, Calin GA, Croce CM, Coukos G, Zhang L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle. 2008;7:2643-2646.
- Smonskey MT, Block AW, Deeb G, Chanan-Khan AA, Bernstein ZP, Miller KC, et al. Monoallelic and biallelic deletions of 13q14. 3 in chronic lymphocytic leukemia: FISH vs. miRNA RT-qPCR detection. Am J Clin Pathol. 2012;137:641-646.
- Arsham MS, Barch MJ, Lawce HJ, editors. The AGT cytogenetics laboratory manual. John Wiley & Sons. 2017.
- Shaffer LG, McGovan-Jordan J, Schmid M. In: An international system for human cytogenetic nomenclature. S Karger Basel. 2016.
- FISH Probe Cataloque. Cambridge, UK: Cytocell; 2014/2015.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta DeltaC(T)) Method. Methods. 2001;25:402-408
- Jansson MD, Lund AH. MicroRNA and cancer. MolOncol.2012;6:590-610.
- Pagona F, De Marinis E, Grignani F, Nervi C. Epigenetic role of miRNA in normal and leukemic hematopoiesis. Epigenomics. 2013;5:539-552.
- Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer cell. 2007;12:215-229.
- Dicker F, Schnittger S, Haferlach T, Kern W, Schoch C. Immunostimulatory oligonucleotide-induced metaphase cytogenetics detect chromosomal aberrations in 80% of CLL patients: a study of 132 CLL cases with correlation to FISH, IgVH status, and CD38 expression. Blood. 2006;108:3152-3160.
- Kotkowska A, Wawrzyniak E, Blonski JZ, Robak T, Korycka-Wolowie A. Chromosomal aberrations in chronic lymphocytic leukemia detected by conventional cytogenetic with DSP30 as a single agent: Comparison with FISH. Leuk Res.2011;35:1032-1038
- Struski S, Gervais C, Helias C, Herbrecht R, Audhuy B, Mauvieux L. Stimulation of B-cell lymphoproliferations with CpG-oligonucleotide DSP30 plus IL-2 is more effective than with TPA to detect clonal abnormalities. Leukemia. 2009;23:617-619.
- Jenderny J, Goldmann C, Thede R, Ebrecht M, Korioth F. Detection of clonal aberrations by cytogenetic analysis after different culture methods and by FISH in 129 patients chronic lymphocytic leukemia. Cytogenet Genome Res. 2014;144:163-168
- Holmes PJ, Peiper SC, Uppa GK,Gong JZ ,Wang ZX,Bajaj R. Efficacy of DSP30-IL-2/TPA for detection of cytogenetic abnormalities in chronic lymphocytic leukemia/small lymphocytic lymphoma. Int J Lab Hematol. 2016;38:483-489.
- Senouci A, Smol T, TricotS,Bakala J,Moulessehoul S,QuilichiniB, et al. Cytogenetic landscape in 1012 newly diagnosed chronic lymphocytic leukemia. Eur J Haematol. 2019;103:607-613.
- Brejcha M, Stoklasová M, Brychtová Y, Panovská A, Štěpanovská K, Vaňková G, et al. Clonal evolution in chronic lymphocytic leukemia detected by fluorescence in situ hybridization and convantional cytogenetic after stimulation with CpG oligonucleotides and interleukin-2: A prospective analysis. Leuk Res. 2014;38: 170-175.
- Athanasiadou A, Stamatopoulos K, Tsezou A, Vadikolia C, Asteriou O, Fassas A, et al. Report of novel chromosomal abnormalities in a series of 130 chronic lymphocytic leukemia patients studied by classic cytogenetic analysis. Leuk Lymphoma. 2006;47:2084-2087.
- De Braekeleer M, Tous C, Guéganic N, Le Bris MJ, Basinko A, Morel F, et al. Immunoglobulin gene translocations in chronic lymphocytic leukemia: a report of 35 patients and review of the literature. Mol Clin Oncol. 2016;4:682-694.
- Nishida Y, Takeuchi K, Tsuda K, Ugai T, Sugihara H, Yamakura M,et al. Acquisition of t(11;14) in a patient with chronic lymphocytic leukemia carrying t(14;19)(q32;q13.1) and +12. Eur J Haematol. 2013;91:179-182.
- SchliemannI, Oschlies I, Nagel I,MurgaPenasEM,Siebert R,Sander B. The t(11;14)(q13;q32)/CCND1-IGH translocation is a recurrent secondary genetic aberration in relapsed chronic lymphocytic leukemia. Leuk Lymphoma. 2016;57:2672-2676.
- Huh YO, Schweighofer CD, Ketterling RP, Knudson RA, Vega F, Kim JE, et al. Chronic lymphocytic leukemia with t (14;19)(q32;q13) is characterized by atypical morphologic and immunophenotypic features and distinctive genetic features. Am J ClinPathol. 2011;135:686-696.
- Glassman AB, Hayes KJ. The value of fluorescence in situ hybridization in the diagnosis and prognosis of chronic lymphocytic leukemia. Cancer Genet Cytogenet. 2005;158:88-91
- Krichevsky S, Bhavsar EB, Furman RR, Niesvizky R, Ritchie EK, Inghirami G, et al. A clinical review of the co-occurrence of myeloproliferative and lymphoproliferative neoplasms. Blood. 2018;132:4285




 The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.
The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.