Phenotypic Detection and Antibiogram of β‑lactamase‑producing Proteus Species in a Tertiary Care Hospital, India
- *Corresponding Author:
- Dr. Pal N
82, Green Nagar, Durgapura, Jaipur, Rajasthan, India.
E-mail: nitapal@yahoo.com
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
Abstract
Background: Proteus species cause a variety of community‑ and hospital‑acquired illnesses. Synthesis of β‑lactamases is the predominant mechanism for resistance to β‑lactam antibiotics. Among the β‑lactamases, extended spectrum β‑lactamases (ESBLs) and AmpC β‑lactamases are the most common. Aim: The objective of this study was to determine the occurrence of ESBL and AmpC β‑lactamases in Proteus species among various clinical isolates at a tertiary care hospital, India. Materials and Methods: This study was done to identify various species of Proteus from clinical samples (n = 3922). Antimicrobial susceptibility was performed by Kirby–Bauer disc diffusion method. ESBL production was detected by modified double‑disc synergy test and indirect modified three‑dimensional tests and AmpC β‑lactamase production by AmpC disc test and modified Hodge test. Results: Proteus species were isolated in 5.4% (101/1876) specimens. Three Proteus species isolated were Proteus mirabilis 62.4% (63/101), Proteus vulgaris 29.7% (30/101), and Proteus penneri 7.9% (8/101). ESBL producers confirmed by both tests were of 88.1% (89/101). Only AmpC β‑lactamase was produced by four isolates. Coproduction of ESBL and AmpC β‑lactamase was observed in 58.4% (52/89) of isolates. Twelve isolates were non‑β‑lactamase producers. Multidrug resistance (MDR) was found in 95.1% (96/101) of isolates, 50.5% (51/101) were possibly extensively drug resistant and none were pan drug resistant. None of the isolates were resistant to piperacillin‑tazobactam. P. penneri isolates exhibited high resistance to most of the antibiotics. Conclusions: A high prevalence of ESBL and AmpC β‑lactamases was found that concurrently showed MDR. Phenotypic methods for the detection of β‑lactamases are easy and simple and can be implemented in routine diagnostic laboratories along with susceptibility testing. These data will assist the clinicians in the management and control of infections.
Keywords
AmpC β-lactamase, Extended spectrum β-lactamase, Extensively drug resistant, Multidrug resistant, Proteus species
Introduction
Proteus species are widespread in the environment and make up part of the normal flora of the human gastrointestinal tract. They cause a variety of community- and hospital-acquired illnesses, including urinary tract, wound, and bloodstream infections.[1] Proteus species are often difficult to eradicate from the host, especially in individuals with complicated ulcers, wounds, and catheterization or functional abnormalities of the urinary tract.[2] Drug resistance has been increasingly reported for this genus, and the predominant mechanism for resistance to β-lactam antibiotics is by the synthesis of β-lactamases. Among the β-lactamases, the production of extended spectrum β-lactamases (ESBLs) and AmpC β-lactamases is most common.[3]
ESBLs are plasmid-mediated β-lactamases that are capable of efficiently hydrolyzing penicillin, narrow and broad-spectrum cephalosporins and monobactams (aztreonam), do not hydrolyze cephamycin or carbapenems (imipenem, meropenem), and are inhibited by β-lactamase inhibitors (e.g., clavulanic acid, sulbactam, and tazobactam).[4] AmpC β-lactamases confer on the bacterium, resistance to penicillins, cephalosporins, cephamycins, and monobactams, and are also resistant to β-lactamase inhibitors. This lack of inhibition by cephamycins and β-lactamase inhibitors differentiates AmpC β-lactamase producers from the ESBL producers.[5]
Plasmids carrying genes for β-lactamases often carry multiple genes for resistance to aminoglycosides, chloramphenicol, quinolones, sulfonamides, tetracycline, and trimethoprim, limiting therapeutic options. Carbapenems are the treatment of choice for serious infections due to ESBL-producing organisms.[6] Since the genes for multiple antibiotic resistance and β-lactamases are transmissible, it is important that β-lactamases should be tested for organisms isolated in hospital and long-term care facility patient populations.
There is a paucity of information on the documentation of β-lactamases producing Proteus among Gram-negative isolates in the northwest region of India. Therefore, a prospective study was undertaken to determine the occurrence of ESBL and AmpC β-lactamases in Proteus species among various clinical isolates at this institute.
Materials and Methods
Specimen collection
A total of 3922 clinical specimens obtained from suspected cases of bacterial infection were received in the Department of Microbiology between February and April 2014. The various clinical specimens received were sputum, pus, urine, cerebrospinal fluid, tracheal swab, endotracheal aspirate, catheter tip, blood, ear swab, vaginal swab, body fluids and tissues. Demographic data (such as age, sex, inpatient and outpatient status) of the patients were recorded.
Culture and identification
Clinical specimens were processed by standard microbiological methods.[7] Proteus species were provisionally diagnosed based on nonlactose-fermenting colonies on MacConkey agar media (Himedia, Vadhani Industrial Estate, LBS Marg, Mumbai, India) with (or without) swarming on blood agar media (Himedia, Vadhani Industrial Estate, LBS Marg, Mumbai, India). Identification of Proteus was done by biochemical tests to find whether they were positive for phenylalanine deaminase production, H2S gas production, citrate utilization, and urease production. Proteus vulgaris produces indole which differentiates it from indole-negative Proteus mirabilis and Proteus penneri. Maltose fermentation and lack of ornithine decarboxylase differentiated P. penneri from P. mirabilis.[7]
Antimicrobial susceptibility test
Kirby–Bauer disc diffusion method was used to test the susceptibility of the Proteus isolates to different antimicrobial agents (Himedia, Vadhani Industrial Estate, LBS Marg, Mumbai, India): amoxicillin-clavulanic acid (20/10 μg), piperacillin-tazobactam (100/10 μg), cefotaxime (30 μg), ceftazidime (30 μg), cefuroxime (30 μg), cefepime (30 μg), cefoxitin (30 μg), amikacin (30 μg), cotrimoxazole (25 μg), gentamicin (10 μg), ciprofloxacin (5 μg), netilmicin (30 μg), imipenem (10 μg), meropenem (10 μg), and doxycycline (30 μg). Escherichia coli American type culture collection (ATCC) 25922 was used as control, and the results were interpreted as per the Clinical Laboratory Standards Institute (CLSI) criteria.[8]
Screening for extended spectrum β-lactamases
As per the CLSI recommendation, isolates showing zone of inhibition ≤22 mm for ceftazidime and ≤25 mm for ceftriaxone by disc diffusion method were considered potential ESBL producers. These isolates were further tested for confirmation by modified double-disc synergy test (MDDST) and indirect modified three-dimensional tests.[4]
Modified double-disc synergy test
Lawn culture of test strain was prepared on Muller Hinton agar (MHA) media (Himedia, Vadhani Industrial Estate, LBS Marg, Mumbai, India), a disc of piperacillin-tazobactam (100/10 μg) was placed at a distance of 25 mm from cefepime (30 μg) disc (center to center). The organisms were considered to be ESBL producers when the zone of inhibition around cefepime showed a distinct increase toward the piperacillin-tazobactam disc [Figure 1].[4]
Indirect modified three-dimensional test
Lawn cultures of ATCC E. coli 25922 were prepared on MHA plate, a disc of ceftriaxone (30 μg) was placed in the center of the plate. A well of 4 mm diameter was punched at a distance of 2 mm from the antibiotic disc. The inoculum (20 μL) of the test strain was adjusted to 5.0 McFarlands standard and was seeded into the well. Heart-shaped distortion of zone of inhibition around the disc was indicative of ESBL production [Figure 2].[4]
Screening for AmpC β-lactamases
Isolates showing a zone of inhibition of <18 mm for cefoxitin by disc diffusion method were considered potential AmpC producers and further confirmed by AmpC disc test and modified Hodge test.
AmpC disc test
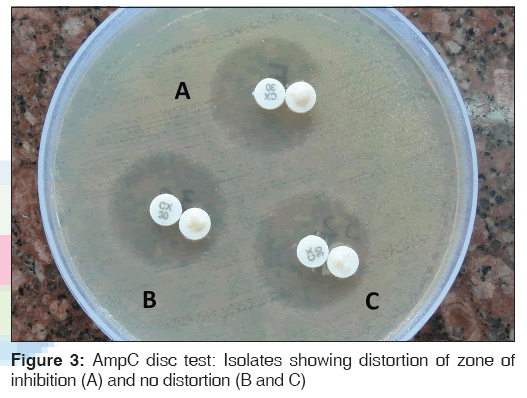
Lawn cultures of ATCC E. coli 25922 were prepared on MHA plate, and a 30 μg cefoxitin disc was placed on the inoculated surface of the agar. A sterile plain disc moistened with sterile saline (20 μL) was inoculated with several colonies of the test organism and was placed beside the cefoxitin disc almost touching it. After overnight incubation at 37°C, the plates were examined for either an indentation or a flattening of the zone of inhibition, indicating enzymatic inactivation of cefoxitin (positive result), or the absence of a distortion, indicating a negative result [Figure 3].[5]
Modified Hodge test
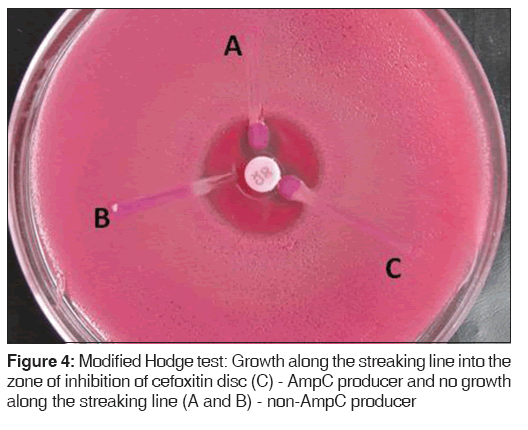
Lawn cultures of ATCC E. coli 25922 were prepared on MacConkey agar plates, cefoxitin disc (30 μg) was placed, and the test organism was streaked toward the cefoxitin disc. If the organism expressed AmpC, it hydrolyzed cefoxitin and showed growth along the intersection of the streak and zone of inhibition of cefoxitin disc [Figure 4].[9]
Quality control
Each batch of the prepared media was checked for sterility for 24 h. The CLSI reference strains of ESBL-positive Klebsiella pneumoniae ATCC 700603 and ESBL-negative E. coli ATCC 25922 were used as controls in the study.
Statistical analysis
Chi-square test was applied for analysis of categorical data. All statistical calculations were performed using MedCalc Statistical Software, version 14.12.0 (MedCalc Software bvba, MedCalc Ostend, Belgium). P < 0.05 was considered statistically significant for interpretation.
Results
A total of 3922 clinical specimens were received for bacteriological culture and antimicrobial susceptibility testing. Microorganisms were isolated in 1876 specimens, out of which 5.4% (101/1876) were Proteus species. Majority of the Proteus species were isolated from pus 80.2% (81/101) followed by urine 8.9% (9/101), vaginal swab 3.0% (3/101), tissue 3.0% (3/101), blood 2.0% (2/101), sputum 2.0% (2/101), and body fluids 1.0% (1/101).
Three Proteus species recovered from 101 specimens were P. mirabilis, 62.4% (63/101); P. vulgaris, 29.7% (30/101); and P. penneri, 7.9% (8/101).
In the present study, Proteus species isolated from inpatients were highest from pus accounting for 67.3% (68/101) of the total isolates followed by urinary isolates accounting for 6.9% (7/101). A total of 80.9% (51/63) P. mirabilis isolates were isolated from inpatients and 19.1% (12/63) from outpatients. P. vulgaris isolates were isolated from 76.6% (23/30) inpatients and 23.3% (7/30) outpatients. All eight (100%) P. penneri isolates were recovered from inpatients.
Detection of extended spectrum β-lactamases
Among the 101 isolates included in the study, 92 and 68 isolates were found to be screened positive for ESBL and AmpC β-lactamases, respectively.
Distribution of β-lactamase producers from various clinical samples among different species is shown in Table 1.
| Samples β-lactamase-producing | Inpatients | Outpatients | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Proteus spp. (n) | ESBL | ESBL + AmpC | Amp C | MDR | XDR | ESBL | ESBL + AmpC | Amp C | MDR | XDR | |
| Pus | Pm (44) | 13 | 23 | 2 | 35 | 26 | 2 | 4 | 0 | 6 | 2 |
| Pv (21) | 6 | 7 | 2 | 15 | 10 | 1 | 5 | 0 | 6 | 0 | |
| Pp (8) | 2 | 5 | 0 | 7 | 6 | 1 | 0 | 0 | 1 | 1 | |
| Urine | Pm (6) | 4 | 1 | 0 | 5 | 0 | 1 | 0 | 0 | 1 | 0 |
| Pv (1) | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Sputum | Pm (2) | 1 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pv (1) | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | |
| Vaginal | Pm (1) | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| swab | Pv (1) | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 |
| Tissue | Pm (2) | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 2 |
| Pv (1) | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Blood | Pm (0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pv (1) | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | |
| Total | 89 | 27 | 40 | 4 | 68 | 44 | 6 | 12 | 0 | 18 | 6 |
Pm: Proteus mirabilis, Pv: Proteus vulgaris, Pp: Proteus penneri, MDR: Multidrug resistant, XDR: Extensively drug resistant, ESBL: Extended spectrum β-lactamase, AmpC: AmpC β-lactamases
Table 1: Distribution of β-lactamase-producing Proteus species isolated from various clinical specimens
i. Detection of ESBL β-lactamases - Out of 92 screen-positive isolates, 85 (92.4%) were confirmed as ESBL producers by both tests. Most of the ESBL β-lactamase producers were isolated from pus, i.e., 69/85 (81.2%)
ii. Detection of AmpC β-lactamases - Out of 68 screen-positive isolates, 56 (82.3%) were confirmed as AmpC β-lactamase producers by both tests. AmpC β-lactamase producers were mostly isolated from pus, i.e., 48/56 (85.7%). Four isolates were pure AmpC β-lactamase producers
iii. Coproduction of ESBL and AmpC β-lactamases - Among the 89 β-lactamase producers, coproduction of ESBL and AmpC β-lactamase was observed in 52/89 (58.4%) isolates. Maximum coproducers were isolated from pus, i.e. 44/52 (84.6%)
iv. Non-β-lactamase producers - Twelve isolates (eight P. mirabilis and four P. vulgaris) were non-β-lactamase producers.
Antimicrobial resistance pattern
Antibiotic susceptibility test was done by Kirby–Bauer disc diffusion method. Multidrug resistance (MDR) was defined as nonsusceptibility to at least one agent in three or more antimicrobial categories, extensively drug resistant (XDR) was defined as nonsusceptibility to at least one agent in all except two or fewer antimicrobial categories (i.e., bacterial isolates remain susceptible to only one or two categories), and pan drug resistance was defined as nonsusceptibility to all agents in all antimicrobial categories. A limited number of antimicrobial agents were tested, so the bacterial isolates were characterized as “possible XDR.”[10]
Antibiotic resistance patterns of Proteus isolates are shown in Table 2. All the three species were 90%–100% resistant to amoxicillin-clavulanic acid, ceftazidime, and netilmicin. All the three species showed a high degree of resistance to 4th generation cephalosporin cefepime (100% in P. mirabilis and P. penneri and 84.6% (22/26) in P. vulgaris). Least resistance was seen with imipenem accounting for 4-8% of isolates. P. penneri isolates exhibited a high resistance to most of the antibiotics followed by P. mirabilis.
| Antimicrobial categories | Antimicrobial agent | Pm (55), n (%) | Pv (26), n (%) | Pp (8), n (%) |
|---|---|---|---|---|
| Aminoglycosides | Amikacin | 41(74.5) | 18(69.2) | 8 (100) |
| Gentamicin | 41(74.5) | 19(73.1) | 8 (100) | |
| Netilmycin | 54(98.2) | 24(92.3) | 8 (100) | |
| Antipseudomonal penicillins + β-lactamase inhibitors | Piperacillin-tazobactam | 0 | 0 | 0 |
| Carbapenems | Meropenem | 23(41.8) | 8 (30.7) | 2 (25) |
| Imipenem | 3(5.4) | 2(7.7) | 1 (12.5) | |
| Nonextended spectrum cephalosporins; 2nd generation | Cefuroxime | 55 (100) | * | * |
| cephalosporins | ||||
| Extended spectrum cephalosporins; 3rd and 4th generation | Ceftazidime | 54(98.2) | 23(88.5) | 8 (100) |
| cephalosporins | Cefotaxime | 34(63.6) | 18(69.2) | 8 (100) |
| Cefepime | 55 (100) | 22(84.6) | 8 (100) | |
| Cephamycins | Cefoxitin | 38(69.1) | 17(65.4) | 8 (100) |
| Fluoroquinolones | Ciprofloxacin | 37(67.3) | 12(46.1) | 7 (87.5) |
| Folate pathway inhibitors | Cotrimoxazole | 50(90.9) | 20(76.9) | 7 (87.5) |
| Penicillins + β-lactamase inhibitors | Amoxicillin-clavulanic acid | 55 (100) | 26 (100) | 8 (100) |
| Tetracyclines | Doxycycline | 49(89.1) | * | * |
Table 2: Antimicrobial resistance pattern of β-lactamase-producing Proteus species
MDR was found in 95.1% (96/101) of isolates and 50.5% (51/101) were possible XDR. Among β-lactamase producers, 96.6% (86/89) were MDR while 56.2% (50/89) were possible XDR. Twelve isolates (eight P. mirabilis, four P. vulgaris) were non-β-lactamase producers, out of which ten 83.3% (10/12) were MDR and one possible XDR.
Discussion
Proteus species are widespread in the environment and make up part of the normal flora of the human gastrointestinal tract. Proteus ranks third as the cause of infections, particularly in hospital-acquired cases.[5]
In the present study, Proteus species were isolated from 5.4% of specimens. Similar prevalence has been reported in studies by Feglo et al.[11] and Leulmi et al.[12] However, some studies have reported a lower prevalence of 1.1%[13] and 3%[14] while other studies have shown a higher prevalence ranging from 14.4% to 28.7%.[15-17] In our study, the highest percentage of Proteus species was isolated from pus specimens (80.2%) followed by urine (8.9%). Similar observation has been reported by other studies whereas some studies have reported isolates more commonly from urine than other clinical specimens.[3,12-14,17,18]
In the present study, three Proteus species (P. mirabilis, P. vulgaris, and P. penneri) were identified. P. mirabilis was the most common (62.4%) among all the isolates which is similar to the findings of other studies.[11,13,15,19] P. mirabilis was isolated in highest number from urine as it has a higher propensity for colonizing the urinary tract due to difference in its pathogenicity.[11]
Indole-negative Proteus species are invariably incorrectly identified as P. mirabilis, missing the isolates of P. penneri which may be nonswarming on the first isolation. All P. penneri isolates were isolated from pus specimens which is similar to other studies[11,19] whereas Kamga et al. in their study isolated 3%, all of which were from urine specimens.
Proteus express virulence factors associated with adherence, motility, immunoavoidance, nutrient acquisition, host damage, biofilm formation, and endotoxicity and therefore behave as opportunistic pathogens in nosocomial infections.[1] We found majority of Proteus infections among the inpatients (81.18%) as compared to outpatients (18.82%) similar to other studies.[11,14,15,19] All strains of P. penneri were isolated from inpatients.
All the three Proteus species displayed high antimicrobial resistance rates to amoxicillin-clavulanic acid, ceftazidime, cefepime, amikacin, gentamicin, and netilmicin. High antimicrobial resistance has also been observed in many studies.[12,16-21] Resistance to carbapenems in the present study was 12.5% which is in accordance with the studies of Datta et al.[21] and Bahashwan and Shafey;[14] however, other studies have reported a very low resistance of 0%–1%.[12,19,22-24] None of the isolates were resistant to piperacillin-tazobactam in our study and that of Tumbarello et al.[19] while Shenoy et al.,[3] Senthamarai et al.,[22] and Rudresh and Nagarathnamma[23] reported a low resistance of 4%–8%.
P. penneri has the ability to cause major infectious diseases and nosocomial outbreaks. P. penneri was isolated from pus specimens of inpatients. All isolates were MDR (100%) while 75% of them were possible XDR. Feglo et al.,[11] Pandey and Tyagi,[13] Kishore,[20] and Senthamarai et al.[22] also reported high MDR in P. penneri isolates.
There is considerable geographical difference in the occurrence of ESBLs within countries, and hospital-to-hospital variability may also be marked. Our study revealed that 88.1% of Proteus species isolated from clinical specimens were β-lactamase producers while others have reported 60%–70%.[3,13,20,23-29] Pure AmpC producers were observed in 5.8% of isolates similar to Feglo and Opoku,[24] while Shenoy et al.[3] and Rudresh and Nagarathnamma[23] reported 11.9% and 15.1%, respectively. Coproduction of ESBL and AmpC was observed in 58.4% of isolates whereas other studies reported lower prevalence.[3,24,21]
MDR observed in β-lactamase producers and in non-β-lactamase producers was 93.3% and 83.3%, respectively, while possible XDR was observed in 56.2% and 16.6% of isolates, respectively. Drug resistance in β-lactamase producers was significantly higher (MDR - P = 0.000019, possible XDR - P = 0.01) than in non-β-lactamase producers. A high degree of co-resistance to cefuroxime, cefepime, amoxicillin-clavulanic acid, ceftazidime, and netilmicin was observed in β-lactamase producers while non-β-lactamase producers were highly resistant to amoxicillin-clavulanic acid and ceftazidime. Imipenem resistance was observed in six β-lactamase producers and in one non-β-lactamase producer which was not statistically significant (P = 0.84). Carbapenem resistance due to porin loss or the presence of β-lactamases capable of hydrolyzing carbapenemases has been documented in ESBL-producing isolates.
The high prevalence of MDR and possible XDR may be due to our institution being a tertiary care referral institute and most of the specimens were received from inpatients who were exposed to previous antibiotics and had undergone some invasive procedures. In the present study, piperacillin-tazobactam, meropenem, and imipenem were found to be most effective.
The present study has certain limitations: specimen number was small, lack of full information on patient’s history (duration of hospitalization, medical prescription, and clinical syndrome), most of the specimens were obtained from inpatients, and ours being a tertiary care hospital, most of the patients were referred cases.
Conclusions
The presence of Proteus in clinical specimens is of great importance, since like other Enterobacteriaceae, they are opportunistic pathogens and may cause morbidity and mortality. In the present study, majority of the Proteus isolates were obtained from pus samples (80.19%). Three Proteus species recovered were P. mirabilis (62.37%), P. vulgaris (29.70%), and P. penneri (7.92%). Majority of the isolates were from inpatients (81.2%). MDR and possible XDR were seen in 93.3% and 56.2% of β-lactamase producers as compared to 83.3% and 16.6% non-β-lactamase producers, respectively. However, all the isolates were susceptible to piperacillin-tazobactam and most of the isolates were susceptible to imipenem and meropenem which are the only options left for the treatment of Proteus infections.
As Proteus species are ubiquitous in the environment, there is a need to maintain proper hygiene standards within hospital surroundings to reduce the incidence of nosocomial infections. Species identification and study of the epidemiology of antimicrobial resistance will assist in the management and control of infections. Therefore, this study is a step towards the generation of data on the prevalence of antimicrobial-resistant pathogens in our institution.
Acknowledgment
We are thankful for the technical assistance given by Mr. RK Yadav and Mr. Raj Kumar.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- O’Hara CM, Brenner FW, Miller JM. Classification, identification, and clinical significance of Proteus, Providencia, and Morganella. Clin Microbiol Rev 2000;13:534-46.
- Yah SC, Eghafona NO, Oranusi S, Abouo AM. Widespread plasmid resistance genes among Proteus species in diabetic wounds of patients in the Ahmadu Bello university teaching hospital (ABUTH) Zaria. Afr J Biotechnol 2007;6:1757-62.
- Shenoy SM, Mohit, Sinha R. Antibiotic sensitivity pattern of clinical isolates of Proteus species with special reference to ESBL and Amp C production. Indian J Appl Res 2013;3:293-4.
- Modi D, Patel D, Patel S, Jain M, Bhatt S, Vegad MM. Comparison of various methods for the detection of extended spectrum beta lactamase in Klebsiella pneumoniae isolated from neonatal Intensive Care Unit, Ahmedabad. Natl J Med Res 2012;2:348-53.
- Singhal S, Mathur T, Khan S, Upadhyay DJ, Chugh S, Gaind R, et al. Evaluation of methods for AmpC beta-lactamase in gramnegative clinical isolates from tertiary care hospitals. Indian J Med Microbiol 2005;23:120-4.
- Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: A clinical update. Clin Microbiol Rev 2005;18:657-86.
- Koneman EW, Allen SD, Janda WM, Schreckenberger PC, Winn WC. Color Atlas and Textbook of Diagnostic Microbiology. 5th ed. Philadelphia, USA: Lippincott-Raven Publishers; 1997.
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty Third Informational Supplement. CLSI document M100- S23. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2013. p. 44-61.
- Vinoth J, Begum ES, Kumar RS, Ramesh S. Phenotypic detection and antibiogram of AmpC beta-lactamases producing tribe Proteeae in a tertiary care hospital. Asian J Pharm Clin Res 2012;5 Suppl 4:180-2.
- Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pan drug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. European society of Clinical Microbiology and Infectious Diseases 2012;18:268-81.
- Feglo PK, Gbedema SY, Quay SN, Adu-Sarkodie Y, Opoku-Okrah C. Occurrence, species distribution and antibiotic resistance of Proteus isolates: A case study at the Komfo Anokye Teaching Hospital (KATH) in Ghana. Int J Pharma Sci Res 2010;1:347-52.
- Leulmi Z, Kandouli C, Benlabed K, Lezzar A, Ilhem Mihoubi I. Prevalence and evaluation of resistance to antibiotics of genera Proteus, Morganella and Providencia isolates in universityhospital of Constantine, Algeria. Int J Adv Res 2014;2:220-7.
- Pandey JK, Tyagi AK. Prevalence of Proteus species in clinical samples, antibiotic sensitivity pattern and ESBL production. Int J Curr Microbiol Appl Sci 2013;2:253-61.
- Bahashwan SA, Shafey HM. Antimicrobial resistance patterns of Proteus isolates from clinical specimens. Eur Sci J 2013;9:188-202.
- Kamga HL, Assob JC, Nsagha DS, Njunda AL, Nde Fon P, Tchape GN. Epidemiologial studies on Proteeae isolates from clinical specimens in the Laquintinie hospital in Douala, Cameroon. Afr J Clin Exp Microbiol 2012;13:112-20.
- Jabur MH, Saedi EA, Trad JK. Isolation of Proteus mirabilis and Proteus vulgaris from different clinical sources and study ofsome virulence factors from Babylon University, college of medicine. Journal of Babylon University/Pure and Applied Sciences 2013;21:43-8.
- Al-Bassam WW, Kazaz AK. The isolation and characterization of Proteus mirabilis from different clinical samples. J Biotechnol Res Cent 2013;7:24-30.
- Reslinski A, Gospodarek E, Mikucka A. Prevalence of multidrug-resistant Proteus spp. strains in clinical specimens and their susceptibility to antibiotics. Med Dosw Mikrobiol 2005;57:175-84.
- Tumbarello M, Trecarichi EM, Fiori B, Losito AR, D’Inzeo T, Campana L, et al. Multidrug-resistant Proteus mirabilis bloodstream infections: Risk factors and outcomes.Antimicrob Agents Chemother 2012;56:3224-31.
- Kishore J. Isolation, identification and characterization of Proteus penneri – A missed rare pathogen. Indian J Med Res2012;135:341-5.
- Datta P, Gupta V, Arora S, Garg S, Chander J. Epidemiology of extended - spectrum ß - lactamase, AmpC, and carbapenemase production in Proteus mirabilis. Jpn J Infect Dis 2014;67:44-6.
- Senthamarai S, Sivasankari S, Anitha C, Kumudavathi MS, Amshavathani SK, Venugopal V, et al. A study on the antibiotic susceptibility pattern of Proteus spp among various samples.
- Rudresh SM, Nagarathnamma T. Extended spectrum β-lactamase producing Enterobacteriaceae and antibiotic co-resistance. Indian J Med Res 2011;133:116-8.
- Feglo P, Opoku S. AmpC beta-lactamase production among Pseudomonas aeruginosa and Proteus mirabilis isolates atthe Komfo Anokye Teaching Hospital, Kumasi, Ghana. J Microbiol Antimicrob 2014;6:13-20.
- Akesola AO, Adeniji TW. Pattern of extended spectrum beta-lactamase production among Proteus species in Western Nigeria. World J Med Sci 2010;5:94-7.
- Nalini K, Sumathi P. Detection of ESBL and cephalosporinase in urinary tract isolates. J Pharm Biomed Sci 2012;21:1-5.
- Hussein AA. Phenotypic detection of extended-spectrum beta lactamase production in Proteus mirabilis isolation from patients with significant bacteriuria in Najaf provina. Qual Manage J 2013;9:149-60.
- Khan MK, Thukral SS, Gaind R. Evaluation of a modified double-disc synergy test for detection of extended spectrum beta-lactamases in AMPC beta-lactamase-producing Proteus mirabilis. Indian J Med Microbiol 2008;26:58-61.
- Kanayama A, Iyoda T, Matsuzaki K, Saika T, Ikeda F, Ishii Y, et al. Rapidly spreading CTX-M type beta-lactamase producing Proteus mirabilis in Japan. Int J Antimicrob Agent 2010;36:340-2.






 The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.
The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.