Validation of a Sars-Cov-2 Nucleocapside N-Protein Detection Test in Saliva
Received: 05-Jul-2021 Accepted Date: Jul 12, 2021 ; Published: 26-Jul-2021
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
An antigenic method for the quantification of SARS-CoV-2 nucleocapside protein (LUMIPULSE SARS-CoV-2 Ag- Fujirebio) both on naso-pharyngeal swab and on saliva has been evaluated on three groups of subjects: sub-intensive care unit hospitalized patients (n=24), patients discharged from this unit (n=22), controls (n=74). The molecular RT-PCR technique was considered the reference method. The cut-off value of 1.04 pg/mL distinguishes sick (hospitalized) from healthy (controls) with sensibility=0.937 and specificity=0.959; area under the ROC curve (0.978); efficiency=0.90. On saliva the qualitative antigenic result (positive if >cut-off) agrees with the qualitative molecular one (k=0.84). Stratifying by groups, in the hospitalized group (with clear prevalence of positives) there is a concordance of the positives of 97%; in the two groups of patients discharged and controls (with clear prevalence of negatives) there is a concordance on the negatives of 91% and 96%, respectively. The qualitative antigenic result on saliva samples is concordant with the molecular qualitative one on the naso-pharyngeal swab (k=0.76).
Keywords
Antigenic; Faculty; Nucleocapside; Hospitalized; Community medicine
Introduction
Validated and accurate laboratory tests for severe acute respiratory syndrome (Coronavirus-2 SARS-CoV-2) are crucial for the management of Corona Virus Disease 2019 (COVID-19) in order to monitor the spread of the disease during the pandemic and to public health interventions including isolation, quarantine and appropriate clinical management of sick individuals. [1]
The diagnostic tools available so far have been based on the detection of viral genes, human antibodies and viral antigens.
The current gold standard for diagnosing infection is Real- Time Reverse Transcriptase Polymerase Chain Reaction (RT-PCR) for qualitative and quantitative detection of viral nucleic acids using samples collected from Nasal- Pharingeal Swabs (NPS).
However, RT-PCR is not lacking in problematic aspects, linked to the complexity of its execution, to the difficulties of procurement and to costs.
Therefore, the clinical laboratory is constantly looking for alternative analytical methods that can support the diagnosis of the infection. The tests for the detection of the SARSCoV- 2 antigen are particularly interesting for their speed and applicability in the analytical phase to common clinical chemistry tools.
But its performance, using NPS as biological material, although characterized by a high specificity, is too low, hovering around 80- 85% of cases, for its application in all clinical settings of the infection.
Recently, several reports highlighted the clinical usefulness of RT-qPCR analysis of saliva specimens.
However, its clinical usefulness remains controversial because the reported diagnostic sensitivity varies widely between 69.2% and 100%, and it has yet to be thoroughly evaluated due to small sample sizes and a lack of detailed clinical information.
Moreover, to our knowledge, saliva has never been used as an analytical sample in antigenic tests.
The purpose of this study was to evaluate the efficacy of the antigen test using saliva samples in comparison to the molecular test on NPS, considered the reference method, within clinically well- defined groups of subjects. We also tested its screening power in a school population with a low prevalence of infection.
Materials and Methods
Ethics statement
The studies involving human participants were reviewed and approved by the Hospital Ethics Committee.
The patients/participants provided their written informed consent to participate in this study.
Population
The study considered a sample of subjects divided as follows:
• 34 patients admitted to the Sub-Intensive Care Unit of Cotugno Hospital (Naples) with a diagnosis of COVID19 (Hospital group)
• 22 patients discharged from the Sub-Intensive Care Unit at the end of the acute phase of COVID19 (Discharge group)
• 74 control subjects recruited from the employees/workers of the Monaldi Hospital for periodic control for molecular swab for SARS-COV-2 (Control group).
At the end of the validation, which was conducted on the first three groups, the test was then performed on a group of 104 students from a high school in Portici (Naples).
Samples
A naso-pharyngeal swab sample (N-) and a Saliva sample (S-) were taken for each subject; each record (sample in the dataset) refers to a sampling of a subject considered only once.
The antigen search was performed within one hour from collection.
The samples were then stored at -80°C until nucleic acid (RNA) extraction.
The most viscous saliva samples were diluted 1:2 with PBS to allow their solubility; the samples with low starting material were diluted up to 1:50 to make the volume acceptable for measurements, subsequently centrifuged at 3,000 rpm for 1 minute with multispinmicrocentrifuge; the analysis was performed on supernatant.
Analytical methods
Method under evaluation (antigenic): the LUMIPULSE SARSCoV- 2 Ag method (Fujirebiocompany) measures the concentration (pg/mL) of the nucleocapside protein SARSCoV- 2 (AgN) in naso- pharyngeal swab and saliva with CLEIA technology (enzyme immunoassay in chemiluminescence) on the LUMIPULSE G600II/ 1200 analyzer (Fujirebio).
Comparison method (PCR): Real Time PCR (RT-PCR) was performed with the BOSPHORE NOVEL CORONAVIRUS kit (2019-Ncov) from ANATOLIA company on BIORAD CFX 96 analyzer. The number of cycles needed to detect the targets (N, ORF1ab, E genes) was used as the unit of measurement.
Statistical analysis
The analytical data were collected in a data set structured as follows in Table:
| material: | Naso-pharyngeal swab | Saliva | ||||||
| method: | In evaluation mAg | reference mPCR | In evaluation mAg | reference mPCR | ||||
| analyte: | T-AgN | T-gORF1AB | T-gN | T-gE | S-AgN | S-gORF 1AB | S-gN | SgE |
Data were statistically evaluated using MedCalc software version 19.6.4- © 2021 MedCalc Software Ltd.
Results
A brief description of the S-AgN results in the three groups of subjects is shown in Table 1.
| HOSPITAL | DISCHARGE | CONTROL | |
|---|---|---|---|
| n | 34 | 22 | 74 |
| average | 175 | 0.51 | 0.24 |
| median | 26.19 | 0.25 | 0.11 |
| minimum | 0.1 | 0.01 | 0.01 |
| maximum | 1987 | 2.33 | 2.1 |
Table 1: Statistical description of the values of S-AgN (pg/mL) in the groups.
A graphical visualization of the distribution of the values in the three groups is shown in Figure 1
Although the differences are all statistically significant, it can be observed that the distribution of the values of the discharge group is more similar to the control group one than to that of hospital one.
Diagnostic features of the antigenic method on saliva.
The first step in evaluating the diagnostic characteristics of a test is to establish a cut-off value beyond which the numerical result must be considered positive.
Typically this value is calculated as the 99th percentile of the distribution of values for a group of normal subjects (URL, Upper Reference Limit).
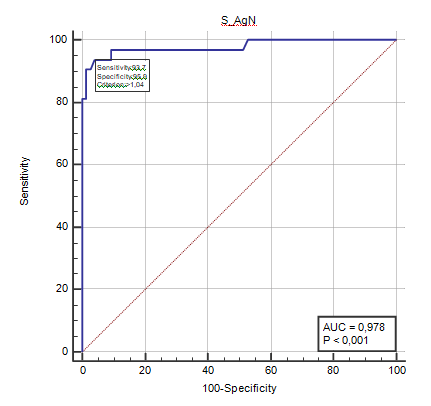
In this case, the 99th percentile in the control sample would be 1.9 pg/mL. Given the high value of this limit (compared to that indicated by the manufacturer: 0.63 pg/mL), we directly considered the complete analysis of the ROC curve, identifying the hospital group as sick and the control group as healthy [Figure 2].
The analysis returns the cut-off value (1.04 pg/mL) at which there is a sensitivity=0.937 and a specificity=0.959 and at which the area under the ROC curve (0.978) and the efficiency (0.90) are maximum.
In an attempt to identify a cut-off also for the group of patients in discharge, a similar ROC analysis was carried out considering the patients in discharge (Discharge) as healthy group and the hospital group as sick group. The obtained results are very similar to the previous analysis (cut-off=0.96; sensitivity=0.937; specificity=0.909; AUC=0.956). The data is consistent with the similarity that the discharge group shows with the control group.
Since the antigen test on naso-pharyngeal swab was not performed in the control (healthy) group, it is not possible to make a ROC analysis on naso-pharyngeal swab material as well.
Comparison between the antigenic method and the molecular method
In order to compare the results of the antigenic method to the molecular method ones, the concordance test (Cohen's k) was used based on the Positive/Negative qualitative judgment of the two methods and the linear correlation test between the numerical values of the antigenic test expressed in pg/mL and those of the PCR method expressed in N° of cycles.
The concordance test between AgN and molecular result (both qualitative, on saliva, on the whole sample of individuals) gave a value of k=0.84 (very good).
Since it cannot be applied the concordance test to each group –due to inconsistency of the data- in order to evaluate the stratified agreement in the three groups admission, discharge and control, it can be simply evaluated the ability of the antigen test to detect positivity/negativity in agreement with the positivity/negativity of the molecular test where the stratification data are consistent [Table 2]:
| Material: Saliva | HOSPITAL | DISCHARGE | CONTROL | % concord | ||
|---|---|---|---|---|---|---|
| n | % concord | n | % concord | n | ||
| Negatives (Antigenic) | * | 20 | 91% | 71 | 96% | |
| Negatives (Molecular) | * | 22 | 74 | |||
| Positives (Antigenic) | 30 | 97% | * | * | ||
| Positives (Molecular) | 31 | * | * |
*: Inconsistent data.
Table 2: Agreement between the antigenic and molecular methods limited to negative cases in the discharge group and control group and to positive cases in the hospital group.
Finally, there is also a good agreement between qualitative S-AgN and swab-qualitative judgment (k=0.76).
Screening application
The saliva auto-sampling was used to carry out a screening by means of both antigen and molecular tests in parallel. 1 out 105 students examined tested positive with molecular test and >99th percentile for S-AgN (0.83 pg/mL) [Table 3].
| S-AgN (pg/mL) | |
|---|---|
| N | 105 |
| average | 0.1 |
| median | 0.06 |
| min | 0.01 |
| max | 0.83 |
| 99° p.le | 0.57 |
Table 3: Results of the antigenic method used as a screening tool for 105 students.
Discussion
By definition, a population screening involves a public health intervention through an early diagnosis tool on asymptomatic subjects. Currently the molecular test on nasopharyngeal swab is the reference diagnostic system for SARS‐CoV-2 infection, due to its high specificity and sensitivity, both around 99%.
However, even the molecular test does not fully satisfy the characteristics of speed, simplicity of execution and cost‐effectiveness necessary for carrying out a mass screening during a pandemic. In fact, the demand for exams often exceeds the laboratories short-term response capacity.
Delay in issuing results delays decisions about isolating infected people, or vice versa forces the temporary isolation of healthy contacts, which in many circumstances can cause personal and financial costs.
Moreover, the correct execution of the NPS is highly dependent on the operator and requires qualified personnel, who are exposed to a high risk of contracting the infection and involves a high organizational effort in the context of a mass screening program.
Saliva contains a pool of microorganisms coming from the lower respiratory tract, nasopharynx, and infected salivary glands, which overexpress ACE2 receptors. In some coronaviruses infection, salivary glands were infected very early in the disease process. Each individual produces about 3,000 droplets of saliva in a single cough, while a sneeze emits approximately 40,000 droplets of saliva with a diameter ranging from 20 to 60 μm, an effect comparable to a person speaking for at least 5 minutes. This makes saliva, in case of infection even in asymptomatic subjects, potentially rich in SARS-Cov2. [2]
In our experience the antigenic method applied on salivary samples presents aspects of extreme interest. With the cut-off value of 1.04 pg/mL, it distinguishes between sick subjects from healthy ones with a specificity and sensitivity respectively of 0.937 and 0.959, an area under the ROC curve of 0.978, an efficiency equal to 0.90 and shows a high level of concordance between its qualitative results and those of the molecular test (k=0.84).
Moreover, stratifying for the different groups studied, a concordance of 97% is reached in that of acute/hospital subjects, which has a clear prevalence of virus-positive cases and of 91% and 96% in the subacute/discharged group and in the control group respectively, which vice versa both have a strong prevalence of virus-negative subjects.
Finally, the qualitative antigenic result on saliva agrees with the qualitative molecular one on NPS (k=0.76) and, most importantly, this analytical approach has been shown to be effective as a screening strategy in a population of students with a low prevalence of infection (1 in 105 schoolchildren). [3]
The World Health Organization (WHO) has published guidelines that validate the use of antigen tests only if equipped with a sensitivity equal to or greater than 80% and a specificity (the ability to give results negative in subjects without infection) equal to or greater than 97% providing for its use not as an alternative to molecular testing, but under particular screening conditions in response to the pandemic. [4]
Our results demonstrate that saliva is better than NPS swab as a material to be analyzed in antigen tests. This analytical approach, in fact, is characterized by equal specificity, but by a higher sensitivity (95% compared to 80-85% of NPS), which makes it a possible alternative to the molecular test due to its lower costs, its speed of execution and its extensibility to a greater number of laboratories, especially in contexts of organizational difficulty, particular pandemic intensity or in countries with weak health systems. [5] But, above all, it can be an easy and quick tool for population screening, especially in schools. Saliva is easily collected by an assisted self- sampling, tilting the head back for 10s and then spit it into a sterile vial. [6] In addition to self‐collection, saliva samples allow performing the test without safe isolation conditions requirement to avoid propagation, thereby widening the settings where the test can be performed, and facilitating the procedure in children since it causes much less discomfort. [7-9]
Conclusion
The antigenic method, applied to the saliva samples of the studied population, has these characteristics: with the cut-off value (1.04 pg/mL) it distinguishes the sick subjects from the healthy ones with sensitivity=0.937 and specificity=0.959; area under the ROC curve (0.978); efficiency=0.90; on saliva the qualitative antigenic result agrees with the qualitative molecular one (k=0.84) stratifying by groups, in the hospital (with a clear prevalence of positives) an agreement between positives of 97% is obtained; in the discharge and control groups (with a clear prevalence of negatives) a concordance on negatives of 91% and 96% is obtained, respectively. The qualitative antigenic result on saliva agrees with the qualitative molecular one on naso- pharyngeal swab (k=0.76).
REFERENCES
- Liu L, Wei Q, Alvarez X, Wang H, Du Y, Zhu H, et al. Epithelial cells lining salivary gland ducts are early target cells of severe acute respiratory syndrome coronavirus infection in the upper respiratory tracts of rhesus macaques. J Virol. 2011;85(8):25–30.
- Xu J, Li Y, Gan F, Du1 Y, Yao Y. Salivary glands: Potential reservoirs for COVID-19 asymptomatic infection. J Dent Res. 2020;99(8):989.
- Xu R, Cui B, Duan X, Zhang P, Zhou X, Yuan Q. Saliva: Potential diagnostic value and transmission ofSARS-CoV-2. Int J Oral Sci. 2020;12(1):1–6.
- World Health Organization. Laboratory testing for corona virus disease 2019 (COVID-19) in suspected human cases: Interim guidance. Geneva, 2020.
- Li H, Liu SM, Yu XH, Tang SL, Tang CK. Coronavirus disease 2019 (COVID-19): Current status and future perspectives. Int J Antimicrobial Agents. 2020;55:10595.
- Di B, Hao W, Gao Y, Wang M, Wang Y, Qiu L, et al. Monoclonal antibody-Based antigen capture enzyme-Linked immunosorbent assay reveals high sensitivity of the Nucleocapsid protein in acute-phase sera of severe acute respiratory syndrome patients. Clin. Diagn. Lab. Immunol. 2005;12 (1):135–140.
- Pang J, Wang M, Ang I, Tan S, Lewis R, Chen J, et al. Potential rapid diagnostics, vaccine and therapeutics for 2019 novel corona virus: A systematic review. J Clin Med. 2020;9(3): 623.
- Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, et al. First case of 2019 novel corona virus in the United States. N Engl J Med. 2020;382(10):929–936.
- Long QX, Deng HJ, Chen YK, Hu Y, Liu BZ, Liao P, et al. Antibody responses to SARS-CoV-2 in COVID-19 Patients: The perspective application of serological tests in clinical practice. Nat Med. 2020;26:845.





 The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.
The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.