BAX and BCL-2 Gene Mutation Signature in Benign and Malignant Lesions of the Breast
2 Department of Medical Laboratory Science, (Histocytopathology Unit) College of Basic and Health Science, Achievers University, Owo, Ondo State, Nigeria
Received: 02-Oct-2022, Manuscript No. AMHSR-22-76406; Editor assigned: 04-Oct-2022, Pre QC No. AMHSR-22-76406 (PQ); Reviewed: 18-Oct-2022 QC No. AMHSR-22-76406; Revised: 20-Feb-2023, Manuscript No. AMHSR-22-76406 (R); Published: 03-Mar-2023, DOI: 10.54608.annalsmedical.2023.86
Citation: Olukayode EV, et al. BAX and BCL-2 Gene Mutation Signature in Benign and Malignant Lesions of the Breast. Ann Med Health Sci Res. 2023;13:451-457
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Background: Apoptosis regulator BAX, also known as bcl-2-like protein 4, is a protein that in humans is encoded by the BAX gene. BAX, a central cell death regulator, is an indispensable gateway to mitochondrial dysfunction and a major pro-apoptotic member of the BCL-2 family proteins that control apoptosis in normal and cancer cells. Dysfunction of apoptosis renders the cancer cell resistant to treatment as well as promotes tumorigenesis. The aim was to study BAX and BCL-2 gene mutations in benign and malignant lesions of the breast.
Materials and methods: A case controlled retrospective study was done with a total of 10 formalin fixed and paraffin wax embedded tissue blocks retrieved from pathology archives, amongst these were 5 benign fibroadenoma and 5 m alignant invasive ductal carcinoma confirmed cases of the breast analyzed using Nucleic acid amplification techniques, the cells was lysed, separated from cellular debris, precipitated, washed with 70% ethanol and eluted, results were presented in SNPs and functional mutation.
Results: This study showed that Bax mutation In Fibroadenoma SNPs observed, 100% transversion, 0% indel and transition. Functional mutation in Fibroadenoma observed, 100% missense, 0% silent and nonsense. In Invasive ductal adenocarcinoma SNPs observed, 58% indel, 29% transversion and 14% transition. Functional mutation in Invasive ductal adenocarcinoma observed, 100% missense and 0% nonsense and silent. BCL-2 mutation in fibroadenoma SNPs observed, 100% transition, 0% indel and 0% transversion. Functional mutation in fibroadenoma observed 67% missense, 0% nonsense and 33% silent. In invasive ductal carcinoma SNPs observed, 30% transversion, 0% insertion and 70% transition. Funcional mutation in invasive ductal carcinoma observed, 40% missense, 60% silent and 0% nonsense. Comparison between BAX and BCL-2 gene in fibroadenoma S NPs observed, 6 0% transition, 4 0% transversion a nd 0% indel. Functional mutation in fibroadenoma observed 8 0% Missense, 20%Silent and 0% Nonsense. In invasive ductal carcinoma SNPs observed, 47% transversion, transition 29% and 24% indel. Funcional mutation in invasive ductal carcinoma observed, 50% missense and 50%Silent and 0% nonsense.
Conclusion: After carrying out this investigation, BAX gene mutation is more defenseless against invasive ductal adenocarcinoma than Fibroadenoma. As a result, mutation in invasive ductal adenocarcinoma in most cases cannot be rectify. In this study, BAX and BCL-2 gene mutation seems to lead to development of breast cancer and affects the disease progression.
Keywords
BAX; BCL-2; FA; IDC; SNPs
Introduction
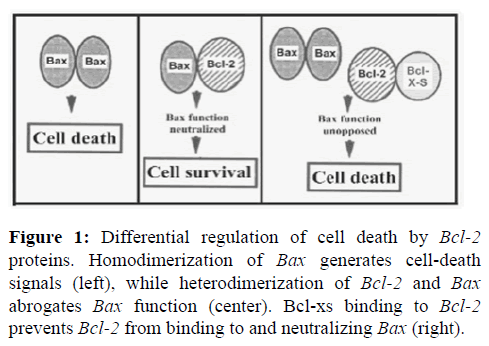
Knowledge apoptosis regulator BAX, also known as BCL-2 like protein 4, is a protein that in humans is encoded by the BAX gene. BAX is a member of the BCL-2 gene family. BCL-2 family members form hetero or homodimers and act as anti or pro-apoptotic regulators that are involved in a wide variety of cellular activities. This protein forms a heterodimer with BCL-2, and functions as an apoptotic activator. This protein is reported to interact with, and increase the opening of, the mitochondrial Voltage Dependent Anion Channel (VDAC), which leads to the loss in membrane potential and the release of cytochrome. The expression of this gene is regulated by the tumor suppressor P53 and has been shown to be involved in P53 mediated apoptosis. BAX, a central cell death regulator, is an indispensable gateway to mitochondrial dysfunction and a major pro-apoptotic member of the Bcl-2 family proteins that control apoptosis in normal and cancer cell. Dysfunction of apoptosis renders the cancer cell resistant to treatment as well as promotes tumorigenesis. BCL-2 protein plays a role in regulating cell death and regulating the balance between new and old cells [1-4].
Cancer and mutation
The cancer genome is moulded by the dual processes of somatic mutation and selection. The selection of advantageous mutations underlies tumorigenesis. The growth of a tumor is therefore a form of evolution at the somatic level, in which the population is comprised of individual cells within the tumor. Models of tumorigenesis have considered the relative importance of mutation and selection. Cancer is a disease of the genome. The classic model of carcinogenesis describes multiple, successive clonal expansions driven by the accumulation of genomic changes or ‘mutations’ that are preferentially selected by the tumour environment. The cancer genome is moulded by the dual processes of somatic mutation and selection. The selection of advantageous mutations underlies tumorigenesis. The growth of a tumor is therefore a form of evolution at the somatic level, in which the population is comprised of individual cells within the tumor. Models of tumorigenesis have considered the relative importance of mutation and selection. Cancer is a disease of the genome. The classic model of carcinogenesis describes multiple, successive clonal expansions driven by the accumulation of genomic changes or ‘mutations’ that are preferentially selected by the tumour environment [5-7].
Materials and Methods
Tissue blocks
In this retrospective study, a total of 10 tissue blocks was selected from the pathology archives of the Federal Teaching Hospital Ido-Ekiti (FETHI). All 10 tissue blocks retrieved were fixed in formalin and embedded tissue blocks consisting of 5 benign fibroadenoma and 5 malignant invasive ductal carcinomas of the breast [8].
DNA extraction
The DNA isolation procedures were similar to those described with minor adjustments to the number of reagents used. The following protocols were adopted for extracting good quality DNA from paraffin embedded tissue. Each tissue was ground in sterile mortal and pistil with 500 μl of extraction buffer before being poured into a sterilized Eppendorf tube and 33 μl of 20% Sodium Dodecyl Sulphate (SDS) was added, vortexed briefly, and incubated in a water bath at 65°C for 10 minutes. 10 μl of 5 M potassium acetate was then added at room temperature, vortexed, and centrifuged at 10000 g for 10 minutes. The supernatant was collected in another Eppendorf tube, and 300 l of cold isopropanol was added. The mixture was gently mixed, and it was kept at -20°C for 60 minutes. The DNA was sediment by centrifugation at 13000 g for 10 minutes, after which the supernatant was gently decanted and the pellet was not disturbed. After washing the DNA pellet with 500 μl of 70% ethanol, it was centrifuged at 10000 g for 10 minutes. Ethanol was decanted, and the DNA was air-dried at room temperature until there was no trace of ethanol left in the tube. To preserve and suspend the DNA, the pellet was suspended in 5 μl of Tris EDTA buffer [9].
Polymerase chain reaction
The cocktail of preparation for PCR sequencing comprised of 10 μl of 5x GoTaq colourless reaction, 3 μl of 25 mM MgCl2, 1 μl of 10 mM dNTPs mix, 1 μl of 10 pmol each primer and 0.3 units of Taq DNA polymerase (Promega, USA) made up to 35 μl with sterile distilled water 15 μl DNA template. PCR was performed in a GeneAmp 9700 PCR System Thermalcycler (Applied Biosystem Inc., USA) with a PCR profile comprising of an initial denaturation for 5 minutes at 94°C, followed by 30 cycles made up of 94°C for 30 seconds, 52°C for 30 seconds and 72°C for 60 seconds; and a final termination for 10 minutes at 72°C [10].
Integrity test
To validate the amplification, the integrity of the amplified DNA of about 1.5 Mb gene fragment was tested on an agarose gel of about 1%. The buffer (1XTAE buffer) was prepared and then used to make the 1.5 percent agarose gel.
The suspension was microwaved for 5 minutes. The molten agarose was allowed to cool to 60°C before being stained with 3 μl of 0.5 g/ml ethidium bromide (which absorbs invisible UV light and transmits the energy as visible orange light). A comb was inserted into the slots of the casting tray, and molten agarose was poured into the tray. The wells were formed by allowing the gel to solidify for 20 minutes. The 1XTAE buffer was poured into the gel tank, barely submerging the gel. After loading the 100 bp DNA ladder into well 1, two microliters (2 μl) of 10X blue gel loading dye (which gives the samples color and density to make it easy to load into the wells and monitor the progress of the gel) was added to 4 μl of each PCR product and loaded into the wells. The gel was electrophoresed at 120 V for 45 minutes, photographed, and then visualized using ultraviolet trans-illumination. The sizes of the PCR products were estimated by comparing them to the mobility of a 100 bp molecular weight ladder that was run in the gel alongside the experimental samples [11].
Purification of amplified product
The amplified fragments were ethanol purified after gel integrity to remove the PCR reagents. In a new sterile 1.5 μl tube Eppendorf, 7.6 μl of Na acetate 3 M and 240 μl of 95% ethanol were added to each about 40 μl PCR amplified product, mix thoroughly by overtaxing, and store at -20°C for at least 30 minutes. Centrifugation for 10 minutes at 13000 g and 4°C, followed by supernatant removal (invert tube on trash once), after which the pellet was washed with 150 μ l of 70% ethanol and mixed, then centrifuged for 15 minutes at 7500 g and 4°C. Remove all supernatant (invert tube on trash) and invert tube on paper tissue and let it dry in the fume hood at room temperature for 10-15 minutes before suspending in 20 μl of sterile distilled water and storing in -20°C prior to sequencing. The purified fragment was tested on a 1.5% agarose gel at a voltage of 110 V for about 1 hour to confirm the presence of the purified product and quantified using a thermos scientific nano drop model 2000 [12].
Sequencing
The amplified fragments were sequenced using an applied bio-systems genetic analyzer 3130 xl sequencer and the Big Dye terminator v3.1 cycle sequencing kit, according to the manufacturer's instructions. For all genetic analyses, the Bio-Edit software and MEGA 6 were used [13].
Data analysis
The results obtained from this study were analyzed using graphs, tables and charts [14].
Results
The effect of mutation shown in the below Table 1.
| Table 1: Summary table showing the effect of mutation along the BAX gene and BCL-2 regions. | |||||
|---|---|---|---|---|---|
| Description | Location | Gene type | % Occurrence | Specimen | Mutation type |
| T | 112 (9:1) | BAX | 1 (11.11) | IDC | Indel |
| G | 70 (9:1) | BAX | 1 (11.11) | IDC | Indel |
| G | 116 (9:1) | BAX | 1 (11.11) | IDC | Indel |
| G | 154 (9:1) | BAX | 1 (11.11) | IDC | Indel |
| C:A | 81 (1:4) | BAX | 1 (11.11) | FA | Transversion |
| C:G | 181 (9:1) | BAX | 1 (11.11) | IDC | Transversion |
| T:G | 119 (9:1) | BAX | 1 (11.11) | FA | Transversion |
| G:C | 121 (9:1) | BAX | 1 (11.11) | IDC | Transversion |
| G:A | 113 (9:1) | BAX | 1 (11.11) | IDC | Transition |
| G:C | 474 (9:1) | BCL-2 | 1 (4) | IDC | Transversion |
| G:T | 780 (4:2) | BCL-2 | 2 (8) | IDC | Transversion |
| G:T | 834 (4:1) | BCL-2 | 2 (8) | IDC | Transversion |
| A:G | 97 (4:1) | BCL-2 | 1 (4) | FA | Transition |
| A:G | 97 (4:1) | BCL-2 | 1 (4) | IDC | Transition |
| G:A | 111 (4:1) | BCL-2 | 2 (8) | IDC | Transition |
| G:A | 306 (3:2) | BCL-2 | 1 (4) | IDC | Transition |
| A:G | 412 (2:3) | BCL-2 | 1 (4) | FA | Transition |
| A:G | 571 (9:1) | BCL-2 | 1 (4) | IDC | Transition |
| A:G | 845 (7:3) | BCL-2 | 3 (12) | IDC | Transition |
| C:T | 843 (4:1) | BCL-2 | 2 (8) | IDC | Transition |
| G:A | 306 (3:2) | BCL-2 | 3 (12) | FA | Transition |
| A:G | 412 (2:3) | BCL-2 | 5 (20) | IDC | Transition |
| Note: This shows the effect of mutation along the BAX and BCL-2 gene. | |||||
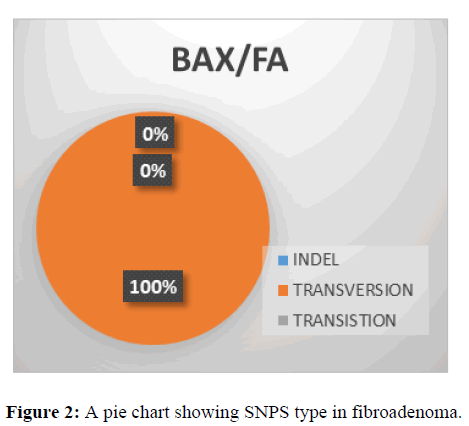
This shows SNPs type in BAX gene, it is observed that the most prevalent mutation is transversion (100%) while indel and transition is 0% (Figure 2).
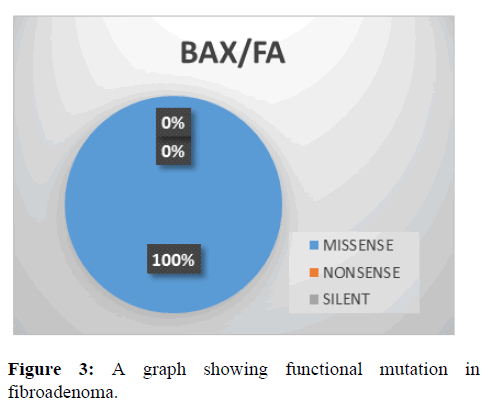
This shows the functional mutation in BAX gene, it is observed that the most prevalent mutation is missense (100%) while nonsense and silent is 0% (Figure 3).
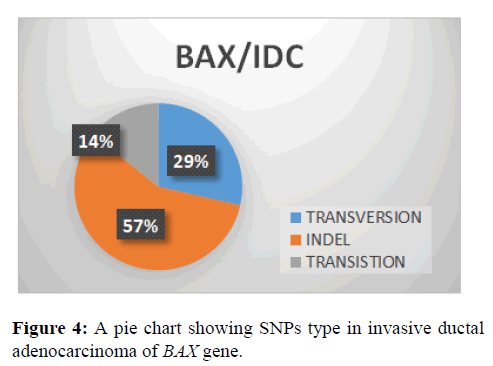
This shows that indel (57%) type of SNPs type in BAX gene followed by transversion with (29%) and transition (14%) (Figure 4).
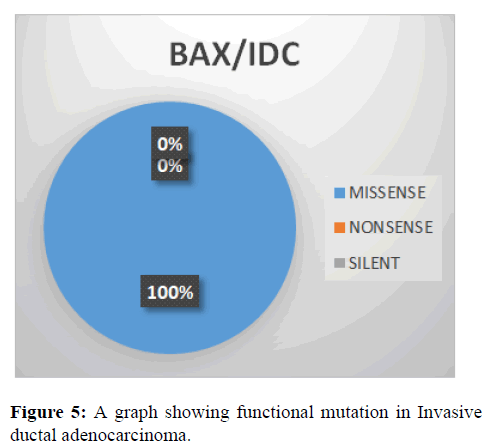
This shows the functional mutation in BAX gene, it is observed that the most prevalent mutation is missense (100%) while nonsense and SILENT is 0% (Figure 5).
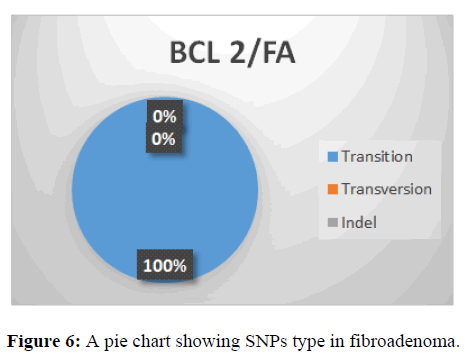
This shows SNPs type in BCL-2 gene, it is observed that the most prevalent mutation is transition (100%) while indel and is transversion 0% (Figure 6).
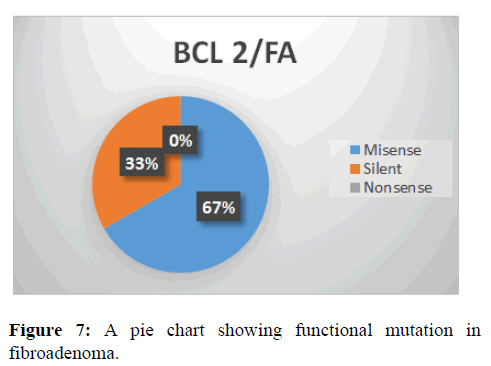
This shows the functional mutation in BCL-2 gene, it is observed that the most prevalent mutation is misense (67%) while nonsense is 0% and silent is 33% (Figure 7).
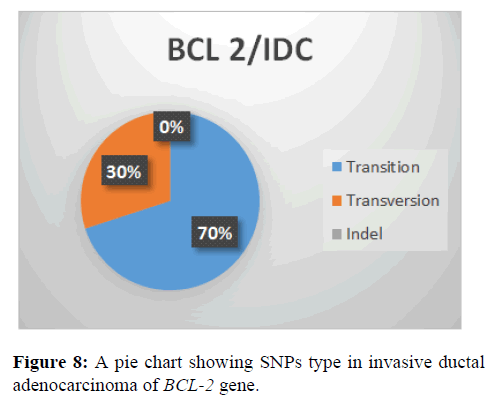
This shows that transition (70%) type of SNPs type in BCL-2 Gene followed by transversion with (30%) and indel (0%) (Figure 8).
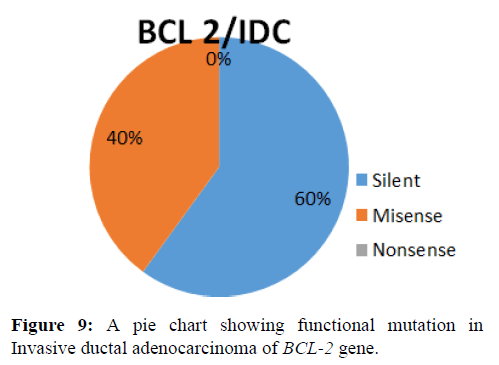
This shows the functional mutation in BCL-2 gene, it is observed that the most prevalent mutation is silent (60%) while misense (40%) and nonsense is 0% (Figure 9).
This shows the functional mutation in BCL-2 gene, it is observed that the most prevalent mutation is silent (60%) while misense (40%) and nonsense is 0% (Figure 10).
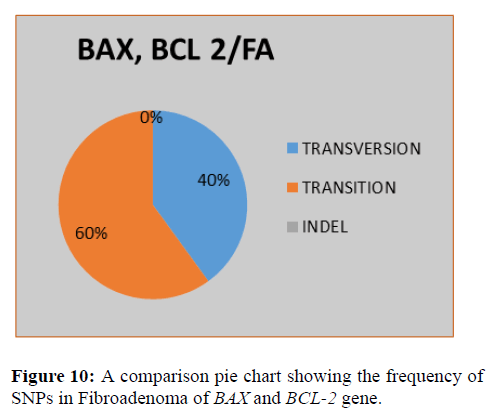
This shows SNPs type in BAX and BCL-2 gene, it is observed that the most prevalent mutation is transition (60%), transversion (40%) and indel (0%) (Figure 11).
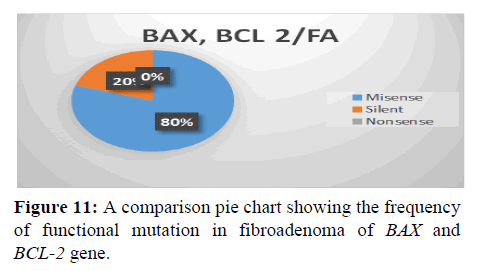
This shows the functional mutation in BAX and BCL-2 gene, it is observed that the most prevalent mutation is missense (80%), silent (20%) and nonsense (0%) (Figure 12).
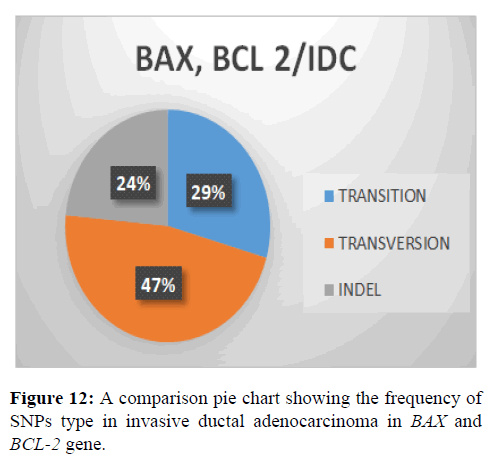
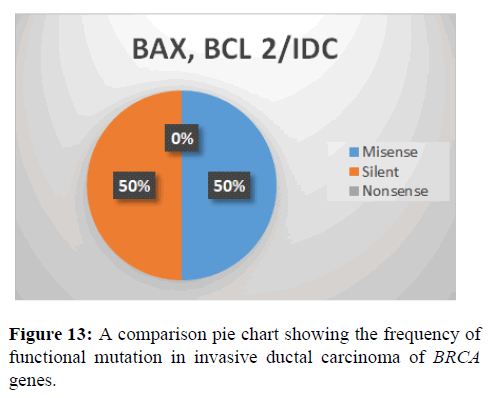
This shows SNPs type in BAX and BCL-2 gene, it is observed that the most prevalent mutation is Transversion (47%), transition (29%) and indel (24%) (Figure 13).
This shows the functional mutation in BAX and BCL-2 Gene, it is observed that the most prevalent mutation is missense (50%), silent (50%) and nonsense (0%).
Discussion
Previous studies have shown that breast cancer is the most common cancer in women worldwide. BAX, as a member of pro-apoptotic gene in BCL-2 family, mainly is a proapoptotic role in the mitochondrial pathway. There have been many studies reporting that Bcl-2, an apoptosis inhibitor, shows a positive correlation with hormone receptors in breast cancer tissues and is a favorable prognostic factor. The imbalance of ant apoptosis and pro-apoptosis gives rise to tumorigenesis. The expression of BAX protein plays an important role in the release of apoptosis-related factors. In this study, In SNPs showed Transition is prevalent. This could indicate spontaneous deamination which aligns with findings. According to Transitions can be caused by oxidative deamination and tautomerization. Although there are twice as many possible transversion, transitions appear more often in genomes, possibly due to the molecular mechanisms that generate them. This study showed Transversion too but not as high as Transition. This could indicate that Transversion has higher regulatory effects than transition. A likely mechanism is that transversion alter the minor groove width and roll of DNA more than transitions, leading to a greater impact on transcription factors binding which aligns with findings. According to transversion are more detrimental because they are more likely to cause substitutions that radically alter biochemical properties of the original amino acid. Indel showed the least occurrence. This indicates that indel are highly deleterious, primarily because two-thirds result in frame-shifts. Bypass through slippage over mononucleotide repeats by transcriptional and translational infidelity is known to occur sporadically which aligns with findings. Indels are supremely important in clinical Next Generation Sequencing (NGS), as they are implicated as the driving mechanism underlying many constitutional and oncologic diseases. In this study, in functional mutation showed equal occurrence in missense and silent. Missense mutation lead to codon changes which leads to production of new protein. According to Loh and Reiter, missense mutation can be related to many pathological conditions and may influence susceptibility to disease and drug treatment. The plausible effects of missense mutations range from affecting the macromolecular stability to perturbing macromolecular interactions and cellular localization. Silent mutations lead to a change of one of the letters in the triplet code that represents a codon, but despite the single base change, the amino acid that is coded for remains unchanged or similar in biochemical properties. According to silent mutations occur when the change of a single DNA nucleotide within a protein-coding portion of a gene does not affect the sequence of amino acids that make up the gene's protein [15-22].
Conclusion
BAX and BCL-2 gene mutation is more defenseless against invasive ductal adenocarcinoma than fibroadenoma. As a result, mutation in invasive ductal adenocarcinoma in most cases cannot be rectify. BAX mutation seems to lead to development of breast cancer and affects the disease progression.
Acknowledgement
All thanks to Almighty God, for his endless mercy and favor.
Conflict of Interest
The authors declare no conflict of interest.
References
- Bergstrom EN, Huang MN, Mahto U, Barnes M, Stratton MR, Rozen SG, et al. Sig Profiler Matrix Generator: a tool for visualizing and exploring patterns of small mutational events. BMC Genomics. 2019; 20:1-12. [Crossref] [Google Scholar] [PubMed]
- Brigatti KW. Introduction to concepts of genetics and genomics. Genomics of Rare Diseases. Cambridge, Academic Press, Massachusetts, USA. 2021; 1-15. [Crossref] [Google Scholar]
- Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J. 2021;134:783-791. [Crossref] [Google Scholar] [PubMed]
- Cannataro VL, Gaffney SG, Townsend JP. Effect sizes of somatic mutations in cancer. J Natl Cancer Inst. 2018;110:1171-1177. [Crossref] [Google Scholar] [PubMed]
- Das A, Sudhaman S, Morgenstern D, Coblentz A, Chung J, Stone SC. et al. Genomic predictors of response to PD-1 inhibition in children with germline DNA replication repair deficiency. Nat Med. 2022; 28:125-135. [Crossref] [Google Scholar] [PubMed]
- Ely B. Genomic GC content drifts downward in most bacterial genomes. Plos one. 2021;16:e0244163. [Crossref] [Google Scholar] [PubMed]
- Jolly C, Van Loo P. Timing somatic events in the evolution of cancer. Genome Biol. 2018;19:1-9. [Crossref] [Google Scholar] [PubMed]
- Kanagavalli U, Deboral E, Lakshmipriya D, Mohamed Sadiq MA, Mohana Priya A. In silico Molecular Docking of Anthraquinone Identified from Boerhavia diffusa Linn against Bax and Bcl-2 Gene. Asia Pac J Sci. 2020;20:175-193. [Google Scholar]
- Katerji M, Duerksen-Hughes PJ. DNA damage in cancer development: special implications in viral oncogenesis. Am J Cancer Res. 2021;11:3956. [Google Scholar]
- Khan A, Kuriachan G, Mahalakshmi R. Cellular Interactome of Mitochondrial Voltage-Dependent Anion Channels: Oligomerization and Channel (Mis) Regulation. ACS Chem Neurosci. 2021;12:3497-3515. [Crossref] [Google Scholar] [PubMed]
- Kunac N, Filipovic N, Kostic S, Vukojevic K. The Expression Pattern of Bcl-2 and Bax in the Tumor and Stromal Cells in Colorectal Carcinoma. Medicina. 2022;58:1135. [Crossref] [Google Scholar] [PubMed]
- Kurschat C, Metz A, Kirschnek S, Hacker G. Importance of Bcl-2-family proteins in murine hematopoietic progenitor and early B cells. Cell Death Dis. 2021;12:1-8. [Crossref] [Google Scholar] [PubMed]
- Liu J, Liu W, Yang H. Balancing apoptosis and autophagy for Parkinson’s disease therapy: targeting BCL-2. ACS Chem Neurosci. 2019;10:792-802. [Crossref] [Google Scholar] [PubMed]
- Loh D, Reiter RJ. Melatonin: regulation of biomolecular condensates in neurodegenerative disorders. Antioxidants (Basel). 2021;10:1483. [Crossref] [Google Scholar] [PubMed]
- Magill ST, Vasudevan HN, Seo K, Villanueva-Meyer JE, Choudhury A, John Liu S, et al. Multiplatform genomic profiling and magnetic resonance imaging identify mechanisms underlying intratumor heterogeneity in meningioma. Nat Commun. 2020;11:1-15. [Crossref] [Google Scholar] [PubMed]
- Nwankwo NJ, KuKui JP, Longdet IY, Kutshik RJ. Polymorphisms in Plasmodium falciparum Dihydropteroate Synthetase (540 pfdhps) Gene Responsible for Sulphadoxine–pyrimethamine Resistant Malaria in Jos, Nigeria. Res Seq. 2022;4:1-16. [Crossref] [Google Scholar]
- Odeyemi AT, Ayantola KJ, Peter S. Molecular Characterization of Bacterial Isolates and Physicochemical Assessment of Well Water Samples from Hostels at Osekita, Iworoko-Ekiti, Ekiti State. Am J Microbiol Res. 2018;6:22-32. [Google Scholar]
- Romero RML, Landerer C, Poehls J, Petroczy AT. Phenotypic mutations contribute to protein diversity and shape protein evolution. Protein Sci. 2022;31:4397. [Crossref] [Google Scholar] [PubMed]
- Singh R, Letai A, Sarosiek K. Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat Rev Mol Cell Biol. 2019;20:175-193. [Crossref] [Google Scholar] [PubMed]
- Skerlev MS, Plavetic ND, Sedlic F, Kuna SK, Vrbanec D, Belev B, et al. Prognostic value of circulating Bcl-2 and anti-p53 antibodies in patients with breast cancer: A long term follow-up (17.5 years). Cancer Biomark. 2021;30:95-104. [Crossref] [Google Scholar] [PubMed]
- Yogosawa S, Yoshida K. Tumor suppressive role for kinases phosphorylating p53 in DNA damage-induced apoptosis. Cancer Sci. 2018;109:3376-3382. [Crossref] [Google Scholar] [PubMed]
- Juba BA, Sovrea A, Crisan D, Melincovici C, Coneac A, et al. Apoptotic markers in photoinduced cutaneous carcinoma. Rom J Morphol Embryol. 2013;54:741-747. [Google Scholar] [PubMed]




 The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.
The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.