Genetically Predicted Sleep Duration in Relation to Risk of Breast Tumor
2 Department of Clinical Oncology, Air force medical University, Shaanxi, China, Email: yajiel_fmmu@foxmail.com
Received: 18-Nov-2024, Manuscript No. amhsr-24-152800 ; Editor assigned: 20-Nov-2024, Pre QC No. amhsr-24-152800 (PQ); Reviewed: 04-Dec-2024 QC No. amhsr-24-152800 ; Revised: 11-Dec-2024, Manuscript No. amhsr-24-152800 (R); Published: 18-Dec-2024
Citation: Lu Y. Genetically Predicted Sleep Duration in Relation to Risk of Breast Tumor. Ann Med Health Sci Res. 2024;S5: 1-7
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Background: Previous observational studies reported that sleep traits were associated with breast tumors. However, the causal relationship between sleep duration and breast tumors is not clear.
Methods: We performed a two-sample bi-directional Mendelian Randomization (MR) analysis with publicly available Genome-Wide Association Studies (GWAS) summary statistics to detect the causal relationship between genetically predicted sleep duration and the risk of breast tumors, including breast cancer (Estrogen Receptor (ER)+/ER-, Human Epidermal Growth Factor Receptor (HER)2+/HER2-), benign breast tumors and carcinoma in situ of the breast. The Inverse Variance Weighting (IVW) method was used as the gold standard for MR results and MR Egger regression, weighted median, weighted mode and simple mode methods were taken as auxiliary analyses. We also conducted sensitivity analyses, including tests for heterogeneity, horizontal pleiotropy and leave-one-out analysis, to evaluate the validity of the detected causal relationships. An additional reverse direction MR analysis was further performed to examine the causal effect of breast tumors on sleep duration.
Design and methods: The study included 311 women aged 20 years to 38 years, who have had at least one pregnancy end in miscarriage and 225 women in the control group. The study of the genotypes of the selected genetic variants was carried out by real-time Polymerase Chain Reaction (PCR) with melting curve analysis.
Results: Suggestive evidence for the causal effect of genetically predicted sleep duration on breast cancer was identified (Odds Ratio (OR)=1.328, 95% Confidence Interval (CI): 1.013–1.741, p=0.04). When distinguished by molecular phenotype, we found a causal association trend between sleep duration and ER+ and ER- breast cancer (ER+ breast cancer p=0.051, ER- breast cancer p=0.077). We did not detect a causal effect of sleep duration on benign breast tumors (OR=1.002, 95% CI: 0.999–1.005, p=0.242) or carcinoma in situ of the breast (OR=1.090, 95% CI: 0.403–2.947, p=0.08). Reverse MR analysis did not indicate that having breast cancer could affect sleep duration (OR = 1.002, 95%CI: 0.995– 1.010, p = 0.494).
Conclusion: The results of this study identified a causative effect of genetically predicted sleep duration on breast cancer, suggesting that ensuring appropriate sleep duration and avoiding excessive sleep might be helpful for preventing breast cancer.
Keywords
Sleep duration; Breast cancer; Mendelian randomization; Causal effect; Prevention
Introduction
Breast cancer is the most common cancer among women worldwide, accounting for a substantial number of new cancer cases and cancer-related deaths [1]. Statistics showed that female breast cancer is the most commonly diagnosed cancer, with an estimated 2.26 million new cases in 2020, surpassing lung cancer as the most prevalent cancer. Understanding the risk factors associated with breast cancer is crucial for prevention efforts. Established risk factors include age, family history of breast cancer, early onset of menstruation, late menopause, reproductive history, hormone replacement therapy, obesity, alcohol consumption and genetic mutations. Lifestyle factors play a crucial role in the development of breast cancer. Smoking has been implicated as a potential risk factor for breast cancer, particularly in premenopausal women, whereas regular physical activity has been associated with a reduced risk of breast cancer [2-5]. Therefore, understanding these modifiable factors can empower individuals to make informed choices and adopt healthy behaviors that may reduce their risk of developing breast cancer.
Sleep traits, such as sleep duration, sleep quality, sleep habits, sleep rhythms, sleep disorders and their relationship to health outcomes have been extensively studied [6,7]. Several studies revealed a relationship between sleep characteristics and the incidence of breast cancer, finding that both short sleep duration and long sleep duration increased the risk of breast cancer [6,8,9]. A study including 23,620 cases found that sleeping less than 6 hours was associated with a 43% increased risk of cancer, including breast cancer [10]. Other studies found a positive association between long sleep duration (>9 hours a night) and breast cancer [11,12]. However, a causal relationship between sleep duration and breast cancer has not been established. Several epidemiological studies and meta-analyses found no association between sleep duration and cancer risk.
Mendelian randomization is a research method that utilizes genetic variants as instrumental variables to establish causal relationships between exposures and outcomes. In Mendelian randomization studies, genetic variants are used as proxies or instrumental variables for modifiable risk factors or exposures of interest. These genetic variants are randomly allocated during meiosis and inherited independently of confounding factors, replicatiom the process of a randomized controlled trial. This random allocation provides a natural experiment-like setting, making Mendelian randomization a powerful tool for causal inference. Compared to other etiological analysis methods, the major advantage of Mendelian randomization is its ability to overcome issues related to confounding and reverse causation, which are common limitations of observational studies. Confounding occurs when an extraneous factor is associated with both the exposure and the outcome, leading to biased estimates of the causal effect. By using genetic variants, which are determined at conception and not influenced by confounders, the risk of confounding bias is greatly reduced. Therefore, Mendelian randomization may provide more valuable analysis into the effects of modifiable risk factors on health outcomes.
In this study, we performed a two-sample MR analysis to investigate whether sleep duration had a causal effect on breast tumors, including breast cancer, ER+/ER- breast cancer, HER2+/ HER2- breast cancer, benign breast tumors and carcinoma in situ of the breast. We also conducted a reverse MR analysis to explore the causal effect of breast tumors on sleep duration. The study aimed to comprehensively and systematically assess the causal relationship between sleep duration and breast tumors and provide evidence for the prevention of breast cancer.
Material and Methods
Study design
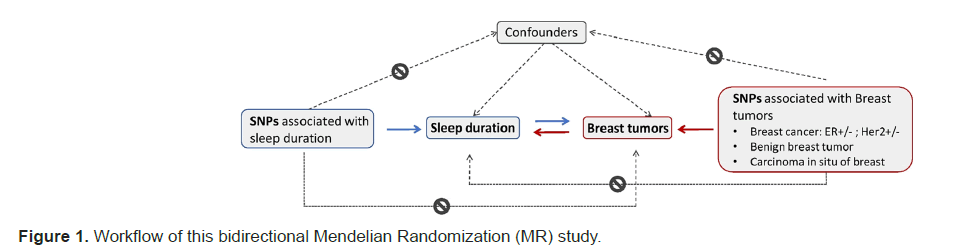
We conducted a bidirectional two-sample MR analysis for each exposure and outcome pair to assess the causal effects between sleep duration and breast tumors. Figure 1 presents the workflow of the study.
Sleep duration and breast tumors were considered exposure factors to assess the causal effects on the other. In bidirectional analysis, Short Nucleotide Polymorphisms (SNPs) related to sleep duration or breast tumors were used as Instrumental Variables (IVs) to deduce causal correlations between exposures and outcomes. MR analysis must meet the following three assumptions to guarantee the reliability of the results: All selected IVs should be strongly related to exposure, no selected IVs should be associated with confounding factors and all selected IVs only influence outcomes through exposure.
Sleep duration data source
Genome-Wide Association Studies (GWAS) datasets were selected from the Independent Evaluation Unit (IEU) OpenGWAS database which contains more than 40,000 GWAS datasets with SNPs related to various traits. The selected sleep duration-related dataset (GWAS ID: ukb-b-4424) consisted of 460,099 samples and 9,851,867 SNPs from European populations published in 2018.
Breast tumor data source
GWAS breast tumor datasets were retrieved from the IEU OpenGWAS database. To further investigate the causal effects of different types of breast tumors, we included GWAS breast cancer datasets (malignant, HER2 and ER status undefined), ER+/ER- breast cancer, HER2+/HER2- breast cancer, benign breast tumors and carcinoma in situ of the breast in MR analysis. Table 1 presents a summary of the data sources of different types of breast tumors, including GWAS ID, number of samples, number of SNPs, population and publication year.
| Trait | GWAS ID | Publication year | Number of samples | Number of SNPs | Population | Source |
|---|---|---|---|---|---|---|
| Breast cancer | ieu-a-1126 | 2017 | 228951 | 10680257 | European | BCAC |
| ER+ breast cancer | ieu-a-1133 | 2017 | 69970 | 10680257 | European | BCAC |
| ER- breast cancer | ieu-a-1166 | 2015 | 21695 | 13011123 | European | BCAC |
| Her2+ breast cancer | finn-b-C3_BREAST_HERPLUS | 2021 | 123302 | 1,63,79,780 | European | BCAC |
| Her2- breast cancer | finn-b-C3_BREAST_HERNEG | 2021 | 119039 | 1,63,79,675 | European | MRC-IEU |
| Benign breast tumor | ukb-b-8549 | 2018 | 4,63,010 | 98,51,867 | European | MRC-IEU |
| Carcinoma in situ of breast | finn-b-CD2_INSITU_BREAST | 2021 | 1,23,579 | 1,63,79,784 | European | MRC-IEU |
Table 1: Data source of different types of breast tumors.
Determination of IVs
The determination of IV was conducted using the “TwoSampleMR” package (version 0.5.7) in R software (version 4.2.2). All SNPs correlated with the exposure trait at a genome-wide significance level (p<5×10−8) were extracted as potential IVs. SNPs in high linkage disequilibrium (r2>0.001 or clump windows < 10,000 kb) were excluded to eliminate bias caused by Linkage Disequilibrium (LD). Harmonization was carried out to eliminate ambiguous SNPs that exhibited non-concordant alleles. When a SNP was not presented in the outcome summary statistics, a proxy SNP highly correlated with the variant of interest (LD, r2>0.8) was selected for substitution. However, if a substitute could not be identified, the SNP was excluded. Palindromic SNPs were aligned when minor allelic frequencies were less than 0.3. IVs with an F-statistic of <10 were excluded due to their weak correlations. To ensure that there was no strong correlation between IVs and confounding factors, we manually removed SNPs that were closely related to confounders after evaluating them using the PhenoScanner tool.
MR statistics
We used the Inverse Variance Weighted (IVW) method to obtain the main results of the two-sample MR analysis. The other four MR analysis methods, including MR Egger regression, weighted median, weighted mode and simple mode methods, were used to obtain secondary results and validation. A clear causal relationship between exposure and outcome was considered for IVW results with P-values of less than 0.05 and the b value of the other four methods was in the same direction as the IVW method b value.
We performed a series of sensitivity analyses, including the heterogeneity test, horizontal pleiotropy test and leave-one-out analysis, to further validate the obtained causal relationships and assess the reliability of the results. The presence of heterogeneity (p<0.05) was allowed in this study. MR-Egger regression was used to investigate the horizontal pleiotropy of the results, whose intercept term indicated possible horizontal pleiotropy. A P-value greater than 0.05 in the horizontal pleiotropy test indicated no horizontal pleiotropy, suggesting no confounding factors. Leave-one-out analysis was employed to evaluate whether the MR results were driven by a single SNP. We performed the same MR statistics in a reverse two-sample MR analysis with breast tumor as the exposure and sleep as the outcome. All MR statistical analyses were conducted using the TwoSampleMR package (version 0.5.7) in R software (version 4.2.2).
Results
Causal effect of sleep duration on breast cancer
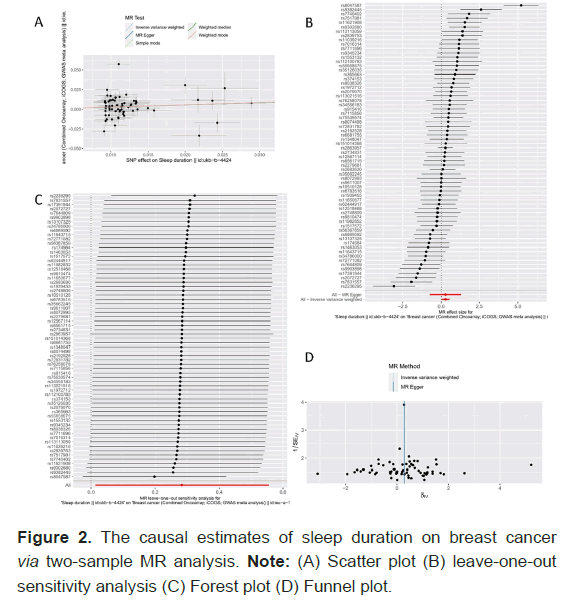
The Figure 2 and Table 2 present the results of the MR analysis of the causal effect estimate of sleep duration on breast cancer, including the evaluation of the pleiotropy effect and heterogeneity. A total of 66 SNPs were finally included in the MR analysis. In the current analysis, we identified a significant causal relationship between sleep duration and the risk of breast cancer, with a p-value of 0.04 (Odds Ratio (OR)=1.328, 95% Confidence Interval (CI): 1.013–1.741) in the IVW analysis (Figure 2B). A positive result was also detected using the weighted median method (OR=1.320, 95% CI: 1.030–1.693, p=0.028). Although no significant results were obtained for other secondary indicators (MR Egger, simple mode and weighted mode methods), the b values were all in the same direction, suggesting a positive association between sleep duration and breast cancer. The MR-Egger regression results confirmed that our results were not significantly affected by horizontal pleiotropy (p=0.995). The funnel plot and Cochran’s Q test indicated significant heterogeneity among the SNPs involved in the IVW analysis (p=1.828e-19) (Figure 2D). A random-effects model was used in the analysis to avoid statistical bias due to heterogeneity. The leave-one-out analysis showed that single SNP removal did not significantly affect the overall estimates (Figure 2C), suggesting that our results were highly reliable.
| Exposure | Outcome | Method | SNPs | OR | 95% CI | b | p-Value | Heterogeneity p-Value (Cochran’s Q Statistic) | MR–Egger Intercept (p-Value) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Sleep duration || ukb-b-4424 | Breast cancer || ieu-a-1126 | MR Egger | 66 | 1.332 | 0.477 | 3.721 | 0.287 | 0.586 | 1.83E-19 | 0.995 |
| Weighted median | 1.32 | 1.03 | 1.693 | 0.278 | 0.028 | |||||
| Inverse variance weighted | 1.328 | 1.013 | 1.741 | 0.284 | 0.04 | |||||
| Simple mode | 1.344 | 0.779 | 2.318 | 0.296 | 0.292 | |||||
| Weighted mode | 1.344 | 0.911 | 1.983 | 0.296 | 0.141 | |||||
| ER+ breast cancer|| ieu-a-1133 | MR Egger | 65 | 1.535 | 0.316 | ~ 7.461 | 0.428 | 0.597 | 2.73E-08 | 0.988 | |
| Weighted median | 1.966 | 1.258 | ~ 3.072 | 0.676 | 0.003 | |||||
| Inverse variance weighted | 1.516 | 0.998 | ~ 2.304 | 0.416 | 0.051 | |||||
| Simple mode | 2.328 | 0.894 | ~ 6.061 | 0.845 | 0.088 | |||||
| Weighted mode | 2.147 | 1.069 | ~ 4.315 | 0.764 | 0.036 | |||||
| ER- Breast cancer|| ieu-a-1166 | MR Egger | 65 | 1.875 | 0.218 | 16.101 | 0.629 | 0.569 | 0.006 | 0.916 | |
| Weighted median | 2.313 | 1.093 | 4.896 | 0.839 | 0.028 | |||||
| Inverse variance weighted | 1.677 | 0.946 | 2.974 | 0.517 | 0.077 | |||||
| Simple mode | 4.011 | 0.827 | 19.452 | 1.389 | 0.09 | |||||
| Weighted mode | 2.724 | 0.848 | 8.746 | 1.002 | 0.097 | |||||
| Breast cancer HER2+|| finn-b-C3_BREAST_HERPLUS | MR Egger | 69 | 0.969 | 0.118 | 7.991 | -0.031 | 0.977 | 0.767 | 0.995 | |
| Weighted median | 1.16 | 0.515 | 2.612 | 0.148 | 0.721 | |||||
| Inverse variance weighted | 0.963 | 0.564 | 1.642 | -0.038 | 0.889 | |||||
| Simple mode | 1.735 | 0.255 | 11.812 | 0.551 | 0.575 | |||||
| Weighted mode | 1.735 | 0.367 | 8.194 | 0.551 | 0.489 | |||||
| Breast cancer HER2-|| finn-b-C3_BREAST_HERNEG | MR Egger | 64 | 0.757 | 0.058 | 9.971 | -0.278 | 0.833 | 0.877 | 0.492 | |
| Weighted median | 1.406 | 0.524 | 3.769 | 0.341 | 0.499 | |||||
| Inverse variance weighted | 1.825 | 0.958 | 3.477 | 0.602 | 0.067 | |||||
| Simple mode | 1.921 | 0.256 | 14.428 | 0.653 | 0.528 | |||||
| Weighted mode | 1.551 | 0.306 | 7.845 | 0.439 | 0.598 | |||||
| Benign neoplasm of breast || ukb-b-8549 | MR Egger | 44 | 0.996 | 0.97 | 1.022 | -0.004 | 0.757 | 0.906 | 0.651 | |
| Weighted median | 1.001 | 0.997 | 1.005 | 0.001 | 0.606 | |||||
| Inverse variance weighted | 1.002 | 0.999 | 1.005 | 0.002 | 0.242 | |||||
| Simple mode | 0.999 | 0.991 | 1.008 | -0.001 | 0.826 | |||||
| Weighted mode | 1 | 0.991 | 1.008 | 0 | 0.949 | |||||
| Carcinoma in situ of breast ||finn-b-CD2_INSITU_BREAST | MR Egger | 69 | 2.061 | 0.041 | 104.478 | 0.723 | 0.719 | 0.811 | 0.743 | |
| Weighted median | 0.96 | 0.214 | 4.315 | -0.041 | 0.958 | |||||
| Inverse variance weighted | 1.09 | 0.403 | 2.947 | 0.086 | 0.865 | |||||
| Simple mode | 0.823 | 0.037 | 18.537 | -0.195 | 0.903 | |||||
| Weighted mode | 1.127 | 0.088 | 14.498 | 0.12 | 0.927 | |||||
Table 2: The results of MR analysis evaluating causal effect of sleep duration on breast tumors.
Causal effect of sleep duration on ER+/ER- or HER2+/ HER2- breast cancer
Additional MR analyses were performed in ER+/ER- and HER2+/HER2- breast cancers to further verify the causal effect between sleep duration and different molecular subtypes of breast cancer. A total of 65 SNPs were finally selected for the causal effect analysis of sleep duration on ER+ breast cancer. Although the IVW method detected no statistical significance, the p-value was close to 0.05 (p=0.051). Analyses using the weighted median and weighted mode methods revealed a significant causal relationship between sleep duration and ER+ breast cancer (weighted median, p=0.003; weighted mode, p=0.036) (Table 2). Similarly, a positive causal effect was identified between sleep duration and ER- breast cancer. A p-value with borderline statistical significance (p=0.077) was obtained using the IVW method. However, a significant statistical difference was obtained by the weighted median method (p=0.028). The results obtained by the other three methods showed similar trends in the causal effect of sleep duration on ER- breast cancer, but without statistical significance (MR Egger p=0.569, simple mode p=0.090, weighted mode p=0.097) (Table 2).
The IVW approach did not show a significant causal connection in the MR analysis of sleep duration on HER2+/HER2- breast cancer (HER2+ breast cancer: OR=0.963, 95%CI: 0.564–1.642, p=0.889; HER2- breast cancer: OR=1.825, 95%CI: 0.958– 3.477, p=0.067) (Table2). Although the p-value for the causal effect of sleep on HER2- breast cancer was close to 0.05, we did not believe that there was a clear causal relationship between sleep duration and HER2- breast cancer because MR Egger's method detected a causal effect in the opposite direction (b=- 0.278).
Causal effect of sleep duration on benign breast tumor
We further analyzed the causal effect of sleep duration on benign breast tumors. A total of 44 SNPs were used for MR analysis. The IVW approach did not show a causal effect of sleep duration on benign breast tumors (OR=1.002, 95%CI: 0.999– 1.005, p=0.242). Secondary analysis methods, including MR- Egger regression (OR=0.996; 95%CI: 0.970–1.022, p=0.757), weighted median (OR=1.001; 95%CI: 0.997–1.005, p=0.606), simple mode (OR=0.999; 95%CI: 0.991–1.008, p=0.826) and weighted mode (OR=1.000; 95%CI: 0.991–1.008, p=0.949) approaches, exhibited the same results (Table 2). The Cochrans Q test results suggested that there was no heterogeneity among the selected SNPs (p=0.906), so the fixed-effects model was adopted for the above analysis. In horizontal pleiotropy, the MR- Egger regression intercept indicated no evidence of pleiotropy (p=0.651). No significant outlier was detected in the leave-one- out analysis, suggesting that the results were robust.
Causal effect of sleep duration on carcinoma in situ of the breast
The causal relationship between sleep duration and carcinoma in situ of the breast was also examined. Heterogeneity was not found for changes in SNPs (p=0.811). The MR analysis found no statistical causal effect using the IVW method (OR=1.090, 95%CI: 0.403–2.947, p=0.086) and the other four methods. Finally, the MR Egger regression intercept provided no evidence for directional pleiotropy (p=0.743) (Table 2).
Reverse MR results
We performed a reverse MR analysis to verify the possibility of reverse causality, in which breast tumors were used as the exposure and sleep duration as the outcome. The IVW method was used to determine whether there was a causal effect of breast tumors on sleep duration. A total of 126 SNPs related to breast cancer were finally included in reverse MR analysis. The IVW analysis results did not indicate that having breast cancer could affect sleep duration (OR=0.998, 95%CI: 0.989–1.006, p=0.670) (Table 3).
| Exposure | Outcome | SNPs | OR | 95% CI | b | p-Value | Heterogeneity p-Value (Cochran’s Q Statistic) | MR–Egger Intercept (p-Value) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Breast cancer || ieu-a-1126 | Sleep duration || ukb-b-4424 | 129 | 1.002 | 0.995 | 1.01 | 0.003 | 0.494 | 1.75E-14 | 0.733 | |
| ER+ breast cancer|| ieu-a-1133 | 38 | 0.998 | 0.989 | 1.006 | -0.002 | 0.67 | 1.88E-10 | 0.683 | ||
| ER- Breast cancer|| ieu-a-1166 | 7 | 1.019 | 0.994 | 1.046 | 0.019 | 0.136 | 1.13E-04 | 0.098 | ||
| Breast cancer HER2+|| finn-b-C3_BREAST_HERPLUS | 8 | 1 | 0.989 | 1.011 | 0 | 0.966 | 0.046 | 0.109 | ||
| Breast cancer HER2-|| finn-b-C3_BREAST_HERNEG | 4 | 0.998 | 0.982 | 1.015 | -0.001 | 0.887 | 0.019 | 0.732 | ||
| Benign neoplasm of breast || ukb-b-8549 | 1 | 2.011 | 0.026 | 153.896 | 0.698 | 0.752 | / | / | ||
| Carcinoma in situ of breast ||finn-b-CD2_INSITU_BREAST | 0 | / | / | / | / | / | / | / | ||
Table 3: The results of MR analysis evaluating causal effect of breast tumors on sleep duration.
Further analysis based on breast cancer molecular typing generated similar results, that ER+/ER- and HER2+/HER2- breast cancer did not have a causal effect on sleep duration. Analysis of the causal relationship between benign breast tumors and sleep duration also did not yield statistically significant results (OR=2.011, 95%CI: 0.26–153.896, p=0.752). However, this analysis included only one SNP and low confidence. No available SNP was identified in the analysis of the causal relationship between carcinoma in situ of the breast and sleep duration. Therefore, there were no results from this part of the reverse analysis.
Discussion
This Mendelian analysis systematically analyzed the causal effect of sleep on breast cancer and its reverse effect. The results suggested evidence for causal effects between sleep duration and the risk of breast cancer. Women with longer sleep duration might have a significantly increased risk of breast cancer. A subgroup analysis based on breast cancer molecular typing revealed a positive correlation trend between sleep duration and ER+/ER- breast cancer. However, we did not detect a clear causal effect of sleep duration on HER2+/HER2- breast cancer, benign breast tumors, or carcinoma in situ of the breast, nor did we find that having breast tumors increased or decreased sleep duration.
There is a traditional belief that shorter sleep duration might be a risk factor for breast cancer and long sleep duration might reduce the risk of cancer. However, our study generated inconsistent conclusions. Sleep duration had a causal effect on breast cancer and longer rather than shorter sleep duration might increase the risk of breast cancer. The positive association between long sleep duration and breast cancer has attracted considerable attention in recent years. A study by Wang et al. reported a positive association between long sleep duration (>9 hours a night) and breast cancer incidence compared to a reference sleep duration (6.1–8.9 hours a night). Another case- control study revealed that the risk of breast cancer increased with increasing sleep duration for every additional sleeping hour (OR=1.06, 95% CI: 1.01–1.11). In addition, a meta-analysis including 10 studies showed that women with longer sleep durations had a significantly increased risk of breast cancer and the effect was dose-dependent. Our study provided definitive evidence for the association between increased breast cancer risk and long sleep duration using the two-sample MR analysis with SNPs as IVs. Unlike prospective or retrospective cohort studies, this analysis eliminated biases that could arise from confounding factors, making the conclusions more reliable.
The detailed mechanism by which sleep duration affects the occurrence of breast cancer is not clear. Several studies suggested that reduced melatonin secretion due to sleep deprivation might be a major mechanism promoting cancer development and melatonin is known to have an effect on protecting the body from cancer. However, such an explanation does not fit our results. The mechanism by which long-term sleep duration increases the risk of breast cancer remains to be analyzed. Long- duration sleepers have higher cortisol levels than short-duration sleepers. Cortisol is involved in multiple processes involved in the genesis and development of breast cancer, such as the regulation of mammary epithelium growth, the impairment of immune activity and the suppression of natural killer cell activity. These effects of cortisol may be the underlying mechanisms by which long-term sleep duration increases the risk of breast cancer. In contrast, high cortisol levels may also promote the growth of ER+ breast cancer by interfering with estrogen signaling pathways. The sub-analysis of this study also detected a causal effect of sleep duration on ER+ breast cancer, which confirmed such a mechanism.
Sleep disorders are common comorbidities in cancer patients. Patients with cancer were reported to be at a high risk of sleep disorders, the most prominent of which was insomnia. No causal effect of breast tumors on sleep duration was detected in this reverse MR analysis. However, we could not draw a strong conclusion that breast tumors do not affect patients’ sleep duration because there were too few SNPs as IVs in the evaluation of ER- breast cancer (SNPs=7), HER2+/HER2- breast cancer (SNPs=8/4), benign neoplasm of the breast (SNPs=1) and carcinoma in situ of the breast (SNPs=0). In fact, the causal effect of breast tumors on sleep duration is complex. Although some patients may experience sleep problems during cancer or treatment, not all patients are affected.
Different types of cancer, as well as different stages of cancer, affect patient’s sleep differently. The prognosis of patients with breast cancer is generally good and endocrine therapy or targeted therapy can replace traditional chemotherapy and reduce toxic side effects.
Therefore, the psychological stress caused by the disease and treatment of breast cancer patients is less than that of other malignant tumors, such as liver cancer and lung cancer. Psychological stress-related anxiety, depression are causes of sleep problems. In contrast, sleep disorders in cancer patients are closely related to treatment factors, individual differences, economic factors and the sleeping environment. Therefore, the sleep problems of breast cancer patients should be paid attention to and this can further improve the quality of life of patients.
This study examined the causal relationship between sleep duration and breast cancer. We utilized genetic variants as IVs to detect the causal relationship between sleep duration and breast tumors, which avoids bias introduced by cofounders. However, some limitations existed in this study. First, heterogeneity was detected in the evaluation of breast cancer, ER+ breast cancer and ER- breast cancer. We used the random-effects IVW method to correct for heterogeneity to ensure the validity of the results. Second, we only used the IEU OpenGWAS database for analysis, which does not represent all GWAS data related to sleep and breast tumors. Finally, sleep traits include sleep duration, chronotype, sleep quality and so on. However, we only analyzed sleep duration in this study. Other sleep factors may be related to breast cancer. Thus, further studies are needed.
Conclusion
In summary, the results of this study suggest the potential causal effect of genetically predicted sleep duration on breast cancer, while a causal effect of sleep duration on benign breast tumors and carcinoma in situ of the breast was not identified. The reverse analysis did not prove the existence of a causal effect of breast tumors on sleep duration. From this study, we recommend that women maintain an appropriate sleep duration and avoid excessive sleep, which is helpful for preventing breast cancer. Large-scale MR studies and clinical trials are required to validate our findings.
Funding
This work was supported by the Youth Talent Promotion Project of Xi'an Association for Science and Technology (No. 959202313008).
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424.
- Macacu A, Autier P, Boniol M, Boyle P. Active and passive smoking and risk of breast cancer: A meta-analysis. Breast Cancer Res Treat. 2015;154:213-224.
- Park HA, Neumeyer S, Michailidou K, Bolla MK, Wang Q, et al. Mendelian randomisation study of smoking exposure in relation to breast cancer risk. Br J Cancer. 2021;125:1135-1145.
- Lynch BM, Neilson HK, Friedenreich CM. Physical activity and breast cancer prevention. Recent Results Cancer Res. 2011:13-42.
- Xu Y, Rogers CJ. Physical activity and breast cancer prevention: Possible role of immune mediators. Front Nutr. 2020;7:557997.
- Richmond RC, Anderson EL, Dashti HS, Jones SE, Lane JM, et al. Investigating causal relations between sleep traits and risk of breast cancer in women: Mendelian randomisation study. BMJ. 2019;365.
- Antza C, Kostopoulos G, Mostafa S, Nirantharakumar K, Tahrani A. The links between sleep duration, obesity and type 2 diabetes mellitus. J Endocrinol. 2022;252:125-141.
- Hery CM, Hale L, Naughton MJ. Contributions of the Women's Health Initiative to understanding associations between sleep duration, insomnia symptoms and sleep-disordered breathing across a range of health outcomes in postmenopausal women. Sleep health. 2020;6(1):48-59.
- Liu L, Bao H, Wang F, Yu L, Cong S, et al. Depressive symptoms and sleep duration as risk factors for breast cancer-china, 2020. China CDC Wkly. 2023;5(15):333.
- Von Ruesten A, Weikert C, Fietze I, Boeing H. Association of sleep duration with chronic diseases in the European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam study. PloS one. 2012;7:e30972.
- Shen J, Chrisman M, Wu X, Chow WH, Zhao H. Sleep duration and risk of cancer in the Mexican American Mano-a-Mano Cohort. Sleep Health. 2019;5(1):78-83.
- Lu C, Sun H, Huang J, Yin S, Hou W, et al. Long‐term sleep duration as a risk factor for breast cancer: evidence from a systematic review and dose‐response meta‐analysis. Biomed Res Int. 2017;2017:4845059.




 The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.
The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.